

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146757.11 - phase: 0 /pseudo
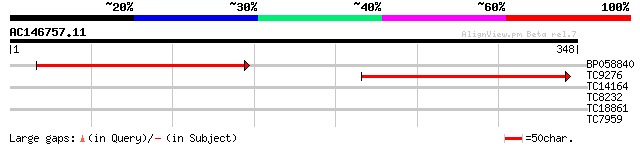
(348 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BP058840 225 1e-59
TC9276 weakly similar to PIR|T00565|T00565 geranylgeranyl-diphos... 176 5e-45
TC14164 weakly similar to UP|Q8VYH6 (Q8VYH6) AT4g17280/dl4675c, ... 28 3.0
TC8232 similar to UP|AAQ57205 (AAQ57205) O-acetylserine (Thiol)l... 27 6.6
TC18861 26 8.7
TC7959 homologue to UP|MDHG_CITLA (P19446) Malate dehydrogenase,... 26 8.7
>BP058840
Length = 464
Score = 225 bits (573), Expect = 1e-59
Identities = 111/131 (84%), Positives = 117/131 (88%)
Frame = +1
Query: 17 KDLHVTFAQMMYHLLPNPYESQEINHLTLAYFVISSLDILNSLHLVEKEAVANWVLSFQV 76
KDLHVTF QMMY LLP+PYESQEINHLTLAYF+IS LDILN+LH VEKEAVAN V SFQV
Sbjct: 70 KDLHVTFLQMMYQLLPSPYESQEINHLTLAYFIISGLDILNALHSVEKEAVANGVXSFQV 249
Query: 77 QRGTTNDPNNGQFYGFHGSKTSQFPPDENGVFHHNNSHLASTYCALAILKIVGYDLSSLD 136
GT DPNNGQFYGFHGS+TSQFPPDENGV HNNSHLAS+YCALAILKIVGYDLSSLD
Sbjct: 250 HPGTKPDPNNGQFYGFHGSRTSQFPPDENGVXLHNNSHLASSYCALAILKIVGYDLSSLD 429
Query: 137 SESMSSSMKNL 147
SE M SSM+NL
Sbjct: 430 SELMLSSMRNL 462
>TC9276 weakly similar to PIR|T00565|T00565 geranylgeranyl-diphosphate
geranylgeranyltransferase I beta chain
homolog At2g39550 - Arabidopsis thaliana {Arabidopsis
thaliana;}, partial (35%)
Length = 607
Score = 176 bits (446), Expect = 5e-45
Identities = 93/128 (72%), Positives = 101/128 (78%)
Frame = +2
Query: 217 LSG*WDTLKTMFSQVVICLL**ICRCCWTGSCRDKGLMVDFKVDLINLLIHVMHFGLEAF 276
LS WD LKT+FSQVV+ LL* +C WTG C +GLMVDFKVDLIN +IHVMHFGLE F
Sbjct: 2 LSDSWDLLKTVFSQVVLRLL*LMCHYYWTGPCSGRGLMVDFKVDLINQVIHVMHFGLELF 181
Query: 277 *EFSGAAILLTIKLYVDFCFPVNTSMVVSANSLETFQIYTIPTMDLQLSACWKNLA*SHF 336
* F GAA L L V+FC P NTSMVVSANSL++FQ YT PTMDL LSACWKNLA*SHF
Sbjct: 182 *GF*GAATL*IRMLSVNFCLPANTSMVVSANSLKSFQTYTTPTMDLVLSACWKNLA*SHF 361
Query: 337 VLNLELLI 344
VLN ELLI
Sbjct: 362 VLNWELLI 385
>TC14164 weakly similar to UP|Q8VYH6 (Q8VYH6) AT4g17280/dl4675c, partial
(44%)
Length = 1606
Score = 27.7 bits (60), Expect = 3.0
Identities = 26/118 (22%), Positives = 48/118 (40%)
Frame = +2
Query: 95 SKTSQFPPDENGVFHHNNSHLASTYCALAILKIVGYDLSSLDSESMSSSMKNLQQPDGSF 154
S T+Q N V N H +L V + + ++ +K + D ++
Sbjct: 686 SGTTQAGSSGNSVRRRRNVH--------GVLNTVSWGILMPLGAVIARYLKVFKSADPAW 841
Query: 155 MPIHIGGETDLRFVYCAGWITGMEWTRKKSRITY*IASLMMVGLD*FLVQNLMVVQLI 212
+H+ +T V AGW TG++ + I Y ++ +G+ F + L V L+
Sbjct: 842 FYLHVTCQTAAYIVGVAGWGTGLKLGSDSAGIEY--STHRALGITLFCLGTLQVFALL 1009
>TC8232 similar to UP|AAQ57205 (AAQ57205) O-acetylserine (Thiol)lyase
(Fragment), partial (42%)
Length = 646
Score = 26.6 bits (57), Expect = 6.6
Identities = 12/35 (34%), Positives = 20/35 (56%)
Frame = +3
Query: 40 INHLTLAYFVISSLDILNSLHLVEKEAVANWVLSF 74
+NHL + Y + +L ++ S LV ++A W L F
Sbjct: 15 LNHLKVQYCLEGNLALIRSKELVLDSSLAFWTLIF 119
>TC18861
Length = 841
Score = 26.2 bits (56), Expect = 8.7
Identities = 9/22 (40%), Positives = 13/22 (58%)
Frame = +1
Query: 166 RFVYCAGWITGMEWTRKKSRIT 187
R ++C GW+TG R+ S T
Sbjct: 481 RIIWCNGWLTGFSRQRRSSEPT 546
>TC7959 homologue to UP|MDHG_CITLA (P19446) Malate dehydrogenase,
glyoxysomal precursor , partial (46%)
Length = 571
Score = 26.2 bits (56), Expect = 8.7
Identities = 18/81 (22%), Positives = 33/81 (40%)
Frame = -2
Query: 12 TTTIDKDLHVTFAQMMYHLLPNPYESQEINHLTLAYFVISSLDILNSLHLVEKEAVANWV 71
T+ D +H + + L + NH T + S D ++++ + WV
Sbjct: 387 TSRYDHQVHASECTLKLRL-----PQKATNHCTSVHVTNISCDTRGIDNIIKMKN*NQWV 223
Query: 72 LSFQVQRGTTNDPNNGQFYGF 92
Q ++G TN P++ Q F
Sbjct: 222 HLHQQRKGLTNSPSSPQDCNF 160
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.335 0.144 0.461
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,566,279
Number of Sequences: 28460
Number of extensions: 92872
Number of successful extensions: 798
Number of sequences better than 10.0: 12
Number of HSP's better than 10.0 without gapping: 792
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 798
length of query: 348
length of database: 4,897,600
effective HSP length: 91
effective length of query: 257
effective length of database: 2,307,740
effective search space: 593089180
effective search space used: 593089180
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.7 bits)
S2: 56 (26.2 bits)
Medicago: description of AC146757.11


