

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
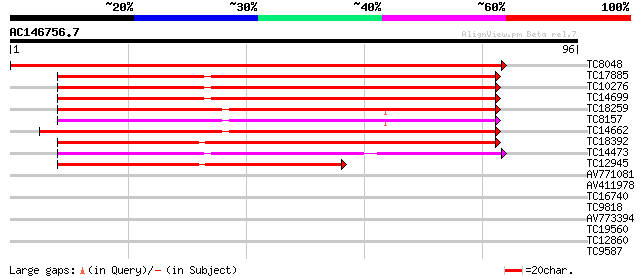
Query= AC146756.7 - phase: 0
(96 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC8048 homologue to UP|PSA6_SOYBN (O48551) Proteasome subunit al... 166 8e-43
TC17885 UP|PSA7_CICAR (Q9SXU1) Proteasome subunit alpha type 7 ... 67 7e-13
TC10276 homologue to UP|PSA7_CICAR (Q9SXU1) Proteasome subunit a... 67 7e-13
TC14699 homologue to UP|PSA5_ORYSA (Q9LSU1) Proteasome subunit a... 65 1e-12
TC18259 homologue to UP|PSA4_SPIOL (P52427) Proteasome subunit a... 64 6e-12
TC8157 homologue to UP|PSA4_SPIOL (P52427) Proteasome subunit al... 62 2e-11
TC14662 similar to UP|PSA3_SPIOL (O24362) Proteasome subunit alp... 61 3e-11
TC18392 homologue to UP|PSA2_ORYSA (Q9LSU2) Proteasome subunit a... 60 8e-11
TC14473 similar to UP|Q8H1Y2 (Q8H1Y2) 20S proteasome alpha 6 sub... 50 8e-08
TC12945 similar to UP|PSA2_ARATH (O23708) Proteasome subunit alp... 46 1e-06
AV771081 27 0.77
AV411978 26 1.00
TC16740 similar to UP|Q9SIJ8 (Q9SIJ8) Expressed protein (RD2 pro... 25 2.2
TC9818 similar to UP|Q7XA50 (Q7XA50) Sorbitol-like transporter, ... 25 2.9
AV773394 24 5.0
TC19560 similar to AAQ56838 (AAQ56838) At1g16900, partial (22%) 23 6.5
TC12860 23 6.5
TC9587 similar to UP|Q9LWB6 (Q9LWB6) 50S ribosomal protein L35, ... 23 8.5
>TC8048 homologue to UP|PSA6_SOYBN (O48551) Proteasome subunit alpha type 6
(20S proteasome alpha subunit A) (20S proteasome
subunit alpha-1) (Proteasome iota subunit) , complete
Length = 1060
Score = 166 bits (419), Expect = 8e-43
Identities = 81/84 (96%), Positives = 83/84 (98%)
Frame = +3
Query: 1 MSRGSGGGYDRHITIFSPEGRLFQVEYAFKAVKAAGITSIGVRGNDSVCVVTQKKVPDKL 60
MSRGSGGGYDRHITIFSPEGRLFQVEYAFKAVKAAGITSIGVRG DS+CVVTQKKVPDKL
Sbjct: 141 MSRGSGGGYDRHITIFSPEGRLFQVEYAFKAVKAAGITSIGVRGKDSICVVTQKKVPDKL 320
Query: 61 LDQTSVTHLFPITKYLGLLATGMT 84
LDQTSV+HLFPITKYLGLLATGMT
Sbjct: 321 LDQTSVSHLFPITKYLGLLATGMT 392
>TC17885 UP|PSA7_CICAR (Q9SXU1) Proteasome subunit alpha type 7 (20S
proteasome alpha subunit D) (20S proteasome subunit
alpha-4) , partial (49%)
Length = 444
Score = 66.6 bits (161), Expect = 7e-13
Identities = 34/75 (45%), Positives = 48/75 (63%)
Frame = +1
Query: 9 YDRHITIFSPEGRLFQVEYAFKAVKAAGITSIGVRGNDSVCVVTQKKVPDKLLDQTSVTH 68
YDR IT+FSP+G LFQVEYA +AV+ G ++GVRG D+V + +KK KL D SV
Sbjct: 88 YDRAITVFSPDGHLFQVEYALEAVR-KGNAAVGVRGTDNVVLGVEKKSTAKLQDSRSVRK 264
Query: 69 LFPITKYLGLLATGM 83
+ + ++ L G+
Sbjct: 265 IVNLDDHIALACAGL 309
>TC10276 homologue to UP|PSA7_CICAR (Q9SXU1) Proteasome subunit alpha type 7
(20S proteasome alpha subunit D) (20S proteasome
subunit alpha-4) , partial (82%)
Length = 688
Score = 66.6 bits (161), Expect = 7e-13
Identities = 34/75 (45%), Positives = 48/75 (63%)
Frame = +2
Query: 9 YDRHITIFSPEGRLFQVEYAFKAVKAAGITSIGVRGNDSVCVVTQKKVPDKLLDQTSVTH 68
YDR IT+FSP+G LFQVEYA +AV+ G ++GVRG D+V + +KK KL D SV
Sbjct: 89 YDRAITVFSPDGHLFQVEYALEAVR-KGNAAVGVRGTDNVVLGVEKKSTAKLQDSRSVRK 265
Query: 69 LFPITKYLGLLATGM 83
+ + ++ L G+
Sbjct: 266 IVNLDDHIALACAGL 310
>TC14699 homologue to UP|PSA5_ORYSA (Q9LSU1) Proteasome subunit alpha type 5
(20S proteasome alpha subunit E) (20S proteasome
subunit alpha-5) , complete
Length = 1044
Score = 65.5 bits (158), Expect = 1e-12
Identities = 31/75 (41%), Positives = 49/75 (65%)
Frame = +1
Query: 9 YDRHITIFSPEGRLFQVEYAFKAVKAAGITSIGVRGNDSVCVVTQKKVPDKLLDQTSVTH 68
YDR + FSPEGRLFQVEYA +A+K G T+IG++ + V + +K++ LL+ +SV
Sbjct: 103 YDRGVNTFSPEGRLFQVEYAIEAIK-LGSTAIGLKTKEGVVLAVEKRITSPLLEPSSVEK 279
Query: 69 LFPITKYLGLLATGM 83
+ I ++G +G+
Sbjct: 280 IMEIDAHIGCAMSGL 324
>TC18259 homologue to UP|PSA4_SPIOL (P52427) Proteasome subunit alpha type 4
(20S proteasome alpha subunit C) (20S proteasome
subunit alpha-3) (Proteasome 27 kDa subunit) , partial
(34%)
Length = 366
Score = 63.5 bits (153), Expect = 6e-12
Identities = 35/76 (46%), Positives = 47/76 (61%), Gaps = 1/76 (1%)
Frame = +2
Query: 9 YDRHITIFSPEGRLFQVEYAFKAVKAAGITSIGVRGNDSVCVVTQKKVPDKLLD-QTSVT 67
YD TIFSPEGRL+QVEYA +A+ AG T+IG+ D V +V +KKV KLL TS
Sbjct: 125 YDSRTTIFSPEGRLYQVEYAMEAIGNAG-TAIGILSKDGVVLVGEKKVTSKLLQTSTSTE 301
Query: 68 HLFPITKYLGLLATGM 83
++ I ++ G+
Sbjct: 302 KMYKIDDHVACAVAGI 349
>TC8157 homologue to UP|PSA4_SPIOL (P52427) Proteasome subunit alpha type 4
(20S proteasome alpha subunit C) (20S proteasome
subunit alpha-3) (Proteasome 27 kDa subunit) , complete
Length = 1080
Score = 61.6 bits (148), Expect = 2e-11
Identities = 34/76 (44%), Positives = 46/76 (59%), Gaps = 1/76 (1%)
Frame = +1
Query: 9 YDRHITIFSPEGRLFQVEYAFKAVKAAGITSIGVRGNDSVCVVTQKKVPDKLLD-QTSVT 67
YD TIFSPEGRL+QVEYA +A+ AG +IG+ D V +V +KKV KLL TS
Sbjct: 145 YDSRTTIFSPEGRLYQVEYAMEAIGNAG-AAIGILSKDGVVLVGEKKVTSKLLQTSTSTE 321
Query: 68 HLFPITKYLGLLATGM 83
++ I ++ G+
Sbjct: 322 KMYKIDDHVACAVAGI 369
>TC14662 similar to UP|PSA3_SPIOL (O24362) Proteasome subunit alpha type 3
(20S proteasome alpha subunit G) (20S proteasome subunit
alpha-7) (Proteasome component C8) , complete
Length = 1110
Score = 61.2 bits (147), Expect = 3e-11
Identities = 29/78 (37%), Positives = 48/78 (61%)
Frame = +2
Query: 6 GGGYDRHITIFSPEGRLFQVEYAFKAVKAAGITSIGVRGNDSVCVVTQKKVPDKLLDQTS 65
G GYD +T FSP+GR+FQ+EYA KAV +G T IG++ D + + +K +P K++ S
Sbjct: 71 GTGYDLSVTTFSPDGRVFQIEYAAKAVDNSG-TVIGIKCKDGIVLGVEKLIPSKMMLPGS 247
Query: 66 VTHLFPITKYLGLLATGM 83
+ + ++ G+ G+
Sbjct: 248 NRRIHSVHRHSGMAVAGL 301
>TC18392 homologue to UP|PSA2_ORYSA (Q9LSU2) Proteasome subunit alpha type 2
(20S proteasome alpha subunit B) (20S proteasome
subunit alpha-2) , partial (71%)
Length = 548
Score = 59.7 bits (143), Expect = 8e-11
Identities = 29/75 (38%), Positives = 49/75 (64%)
Frame = +1
Query: 9 YDRHITIFSPEGRLFQVEYAFKAVKAAGITSIGVRGNDSVCVVTQKKVPDKLLDQTSVTH 68
Y +T FSP G+L Q+E+A AV +G TS+G++ + V + T+KK+P L+D+ SV
Sbjct: 58 YSFSLTTFSPSGKLVQIEHALTAV-GSGQTSLGIKAANGVVIATEKKLPSILVDEASVQK 234
Query: 69 LFPITKYLGLLATGM 83
+ +T +G++ +GM
Sbjct: 235 IQLLTPNIGVVYSGM 279
>TC14473 similar to UP|Q8H1Y2 (Q8H1Y2) 20S proteasome alpha 6 subunit,
partial (86%)
Length = 1302
Score = 49.7 bits (117), Expect = 8e-08
Identities = 28/76 (36%), Positives = 42/76 (54%)
Frame = +3
Query: 9 YDRHITIFSPEGRLFQVEYAFKAVKAAGITSIGVRGNDSVCVVTQKKVPDKLLDQTSVTH 68
YD +T +SP GRLFQVEYA +AVK G +IG+R V + K +L +
Sbjct: 153 YDTDVTTWSPAGRLFQVEYAMEAVK-QGSAAIGLRSKTHVVLACVNKANSEL--SSHQKK 323
Query: 69 LFPITKYLGLLATGMT 84
+F + ++G+ G+T
Sbjct: 324 IFKVDDHIGVAIAGLT 371
>TC12945 similar to UP|PSA2_ARATH (O23708) Proteasome subunit alpha type 2
(20S proteasome alpha subunit B) , partial (93%)
Length = 744
Score = 45.8 bits (107), Expect = 1e-06
Identities = 21/49 (42%), Positives = 33/49 (66%)
Frame = +2
Query: 9 YDRHITIFSPEGRLFQVEYAFKAVKAAGITSIGVRGNDSVCVVTQKKVP 57
Y +T FSP G+L Q+E+A AV +G TS+G++ + V + T+KK+P
Sbjct: 71 YSFSLTTFSPSGKLVQIEHALTAV-GSGQTSLGIKAANGVVIATEKKLP 214
>AV771081
Length = 563
Score = 26.6 bits (57), Expect = 0.77
Identities = 12/26 (46%), Positives = 17/26 (65%)
Frame = +3
Query: 65 SVTHLFPITKYLGLLATGMTGVSFIA 90
SV ++ P+TK +GL T TG+ IA
Sbjct: 63 SVLYIIPVTKQIGLNETQWTGIVRIA 140
>AV411978
Length = 405
Score = 26.2 bits (56), Expect = 1.00
Identities = 11/22 (50%), Positives = 14/22 (63%)
Frame = +2
Query: 57 PDKLLDQTSVTHLFPITKYLGL 78
P KL ++T HLFP+T L L
Sbjct: 65 PSKLTEETHPIHLFPLTSILML 130
>TC16740 similar to UP|Q9SIJ8 (Q9SIJ8) Expressed protein (RD2 protein),
partial (90%)
Length = 765
Score = 25.0 bits (53), Expect = 2.2
Identities = 12/28 (42%), Positives = 17/28 (59%)
Frame = -2
Query: 69 LFPITKYLGLLATGMTGVSFIASPSFLL 96
L PIT GL+ + ++F ASPS +L
Sbjct: 470 LVPITTAAGLILSASLQITFPASPSTIL 387
>TC9818 similar to UP|Q7XA50 (Q7XA50) Sorbitol-like transporter, partial
(62%)
Length = 1162
Score = 24.6 bits (52), Expect = 2.9
Identities = 19/81 (23%), Positives = 34/81 (41%), Gaps = 6/81 (7%)
Frame = +2
Query: 11 RHITIFSPEGRLFQVEYAFKAVKAAGITS------IGVRGNDSVCVVTQKKVPDKLLDQT 64
+++ F P+ + + +YAF A +TS IGV ++ + KV D
Sbjct: 185 KNLQDFDPQKKPKRNKYAFACAMLASMTSILLGYDIGVMSGAAIYIKRDLKVSD-----V 349
Query: 65 SVTHLFPITKYLGLLATGMTG 85
+ L I L+ +G+ G
Sbjct: 350 KIEILLGIINLYSLIGSGLAG 412
>AV773394
Length = 447
Score = 23.9 bits (50), Expect = 5.0
Identities = 10/19 (52%), Positives = 13/19 (67%)
Frame = +2
Query: 37 ITSIGVRGNDSVCVVTQKK 55
ITSI + G +VC VT+ K
Sbjct: 137 ITSIPIEGEGAVCPVTKSK 193
>TC19560 similar to AAQ56838 (AAQ56838) At1g16900, partial (22%)
Length = 516
Score = 23.5 bits (49), Expect = 6.5
Identities = 13/31 (41%), Positives = 17/31 (53%)
Frame = +3
Query: 49 CVVTQKKVPDKLLDQTSVTHLFPITKYLGLL 79
C+ +K LLD SV LFP+ +L LL
Sbjct: 375 CLERKK*ECSMLLDSFSVFSLFPLRLFLWLL 467
>TC12860
Length = 500
Score = 23.5 bits (49), Expect = 6.5
Identities = 9/25 (36%), Positives = 14/25 (56%)
Frame = +1
Query: 68 HLFPITKYLGLLATGMTGVSFIASP 92
H+FP + YLGL+ + +A P
Sbjct: 259 HIFPESAYLGLITD*KCSIPLLAFP 333
>TC9587 similar to UP|Q9LWB6 (Q9LWB6) 50S ribosomal protein L35, partial
(57%)
Length = 819
Score = 23.1 bits (48), Expect = 8.5
Identities = 11/36 (30%), Positives = 18/36 (49%)
Frame = -3
Query: 59 KLLDQTSVTHLFPITKYLGLLATGMTGVSFIASPSF 94
++L +S TH FP T + L + F+A S+
Sbjct: 394 QMLLSSSPTHDFPFTSHAKSLCRRLVSFHFVAFSSY 287
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.322 0.139 0.400
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,297,305
Number of Sequences: 28460
Number of extensions: 12456
Number of successful extensions: 58
Number of sequences better than 10.0: 36
Number of HSP's better than 10.0 without gapping: 56
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 56
length of query: 96
length of database: 4,897,600
effective HSP length: 72
effective length of query: 24
effective length of database: 2,848,480
effective search space: 68363520
effective search space used: 68363520
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 47 (22.7 bits)
Medicago: description of AC146756.7


