

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146755.5 - phase: 0
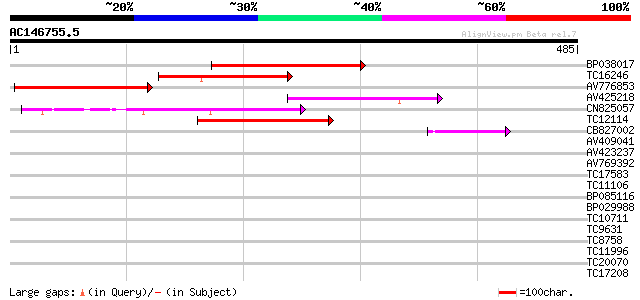
(485 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BP038017 110 7e-25
TC16246 weakly similar to UP|Q8S2M2 (Q8S2M2) Far-red impaired re... 107 4e-24
AV776853 105 1e-23
AV425218 98 3e-21
CN825057 88 3e-18
TC12114 weakly similar to GB|BAC16510.1|23237937|AP005198 transp... 88 4e-18
CB827002 46 2e-05
AV409041 39 0.002
AV423237 35 0.036
AV769392 29 1.5
TC17583 weakly similar to UP|Q9LIE5 (Q9LIE5) Far-red impaired re... 29 1.5
TC11106 29 2.0
BP085116 28 4.4
BP029988 28 4.4
TC10711 27 5.7
TC9631 similar to GB|AAC19270.1|3193286|T14P8 coded for by A. th... 27 5.7
TC8758 similar to GB|AAC77862.2|20197419|AC005623 Argonaute (AGO... 27 7.4
TC11996 similar to GB|AAO63323.1|28950799|BT005259 At2g03430 {Ar... 27 7.4
TC20070 similar to PIR|JQ1073|JQ1073 tryptophan synthase beta-2... 27 9.7
TC17208 similar to UP|Q9MAV5 (Q9MAV5) F24O1.4, partial (13%) 27 9.7
>BP038017
Length = 570
Score = 110 bits (274), Expect = 7e-25
Identities = 47/132 (35%), Positives = 81/132 (60%)
Frame = +2
Query: 173 DAEQAVNFLKHMSSKDDMMFWRHTVHADGSLQHLFWCDGVSCMDYSIFGDVLAFDATYKK 232
DA+ +N+ K M K+ ++ + + + ++FW D S Y+ FGD + FD Y+
Sbjct: 167 DAQNLLNYFKKMQGKNPGFYYAIQLDDENRMINVFWADARSRSAYNYFGDAVIFDTMYRP 346
Query: 233 IKYNTPLVIFSGVNHHNQSIIFGSAIIGDETEDTYVWLLKIFVEAMGGKLPVSVITDGDL 292
+Y P F+GVNHH Q+++FG A++ DE+E ++ WL + ++ AM + PVS+ TD D
Sbjct: 347 NQYQVPFAPFTGVNHHGQNVLFGCALLLDESESSFTWLFRTWLSAMNDRPPVSITTDQDR 526
Query: 293 SMRNAIRKVFPE 304
+++ A+ +VFPE
Sbjct: 527 AIQAAVAQVFPE 562
>TC16246 weakly similar to UP|Q8S2M2 (Q8S2M2) Far-red impaired response
protein-like, partial (4%)
Length = 660
Score = 107 bits (267), Expect = 4e-24
Identities = 51/117 (43%), Positives = 74/117 (62%), Gaps = 2/117 (1%)
Frame = +2
Query: 128 MRQSGISTTRIHGYFASQAGGYQNVGYNRRDMYNE--QRKRRMRWNSDAEQAVNFLKHMS 185
M G+ I GG++++G+ + D+ N + KR N DA A+++L+ +
Sbjct: 8 MNLYGVRACHIMALMLGPKGGHESLGFTKTDLSNHIAKPKRERIQNGDAAAALSYLEGKA 187
Query: 186 SKDDMMFWRHTVHADGSLQHLFWCDGVSCMDYSIFGDVLAFDATYKKIKYNTPLVIF 242
D M F++ T D SL++LFWCDGVS MDY++FGDV+AFD+TYKK KYN PLV+F
Sbjct: 188 DNDPMFFYKFTKTGDESLENLFWCDGVSRMDYNVFGDVIAFDSTYKKNKYNKPLVVF 358
>AV776853
Length = 625
Score = 105 bits (263), Expect = 1e-23
Identities = 45/118 (38%), Positives = 71/118 (60%)
Frame = +1
Query: 5 FPDREVAFMFYNWYGCFHGFAARKSRLIRNINGEVVQQTFLCHREGIREEKYINSTSRKR 64
F E A+ FY Y + GF RK + R+ +G V + F+C+R+G+R +K+ N T RKR
Sbjct: 244 FGSEEEAYQFYQAYAKYQGFIVRKDDIGRDYHGNVNMRQFVCNRQGLRSKKHYNRTDRKR 423
Query: 65 EHKPLSRCGCQAKVRVHIDVSSQRWYIKLFDDDHNHSFVKEKFERMLPAHRKMSEYDK 122
+HKP++ C AK+RVH+D +W + F++ HNH +F +P +R M++ DK
Sbjct: 424 DHKPVTHTNCLAKLRVHLDYKIGKWKVVSFEECHNHELTPARFVHFIPPYRVMNDADK 597
>AV425218
Length = 419
Score = 98.2 bits (243), Expect = 3e-21
Identities = 44/137 (32%), Positives = 75/137 (54%), Gaps = 4/137 (2%)
Frame = +2
Query: 238 PLVIFSGVNHHNQSIIFGSAIIGDETEDTYVWLLKIFVEAMGGKLPVSVITDGDLSMRNA 297
PL GVNHH QS++FG A++ E +++VWL + + M G P +ITD +M+ A
Sbjct: 5 PLATLVGVNHHGQSVLFGCALLSSEDSESFVWLFQSLLHCMSGVPPQGIITDHSEAMKKA 184
Query: 298 IRKVFPEAHHRLCAWHLIRNATSNIKNLHFVSKFK----DCLLGDVDVDVFQRKWEELVT 353
I V P HR C ++++ + + + + V +D F+R W+++V
Sbjct: 185 IETVLPSTRHRWCLSYIMKKLPQKLLGYAQYESIRHHLQNVVYDAVVIDEFERNWKKIVE 364
Query: 354 EFGLEENPWMLEMYQKR 370
+FGLE+N W+ E++ +R
Sbjct: 365 DFGLEDNEWLNELFLER 415
>CN825057
Length = 721
Score = 88.2 bits (217), Expect = 3e-18
Identities = 67/254 (26%), Positives = 111/254 (43%), Gaps = 11/254 (4%)
Frame = +1
Query: 11 AFMFYNWYGCFHGFAA--RKSRLIRNINGEVVQQTFLCHREGIREEKYINSTSRKREHKP 68
A+ FY Y GF + SR + E + F C R G+ E S R
Sbjct: 1 AYSFYQEYAKSMGFTTSIKNSRRSKKTK-EFIDAKFACSRYGVTPESDGGSNRRSS---- 165
Query: 69 LSRCGCQAKVRVHIDVSSQRWYIKLFDDDHNHSFVKEKFERMLPA-------HRKMSEYD 121
+ + C+A + V +W I H F+KE +LPA HR + +
Sbjct: 166 VKKTDCKACMHVKRKPDG-KWII--------HEFIKEHNHELLPALAYHFRIHRNVKLAE 318
Query: 122 KYQMNTMRQSGISTTRIHGYFASQAGGYQNVGYNRRDMYNEQRKRRMRW--NSDAEQAVN 179
K M+ + T +++ + Q+GG N+ D+ ++ +K + DA+ +
Sbjct: 319 KNNMDILHAVSERTRKMYVEMSRQSGGCLNIESLVGDLNDQFKKGQYLAMDEGDAQVMLE 498
Query: 180 FLKHMSSKDDMMFWRHTVHADGSLQHLFWCDGVSCMDYSIFGDVLAFDATYKKIKYNTPL 239
+ KH+ ++ F+ ++ + L+++FW D S DY F DV++FD +Y K P
Sbjct: 499 YFKHIQKENPNFFYSIDLNEEQRLRNIFWIDAKSINDYLSFNDVVSFDTSYIKSNEKLPF 678
Query: 240 VIFSGVNHHNQSII 253
F GVNHH Q I+
Sbjct: 679 APFVGVNHHCQPIL 720
>TC12114 weakly similar to GB|BAC16510.1|23237937|AP005198 transposase-like
{Oryza sativa (japonica cultivar-group);}, partial (8%)
Length = 488
Score = 87.8 bits (216), Expect = 4e-18
Identities = 37/117 (31%), Positives = 71/117 (60%)
Frame = +1
Query: 161 NEQRKRRMRWNSDAEQAVNFLKHMSSKDDMMFWRHTVHADGSLQHLFWCDGVSCMDYSIF 220
+E+R++R + +A + + + ++ + + + + + ++FW D +DY F
Sbjct: 7 SEKRRQRSLLHGEAGYILQYFQRKLVENSPFYHAYQLDDEDQITNVFWVDARMLIDYGYF 186
Query: 221 GDVLAFDATYKKIKYNTPLVIFSGVNHHNQSIIFGSAIIGDETEDTYVWLLKIFVEA 277
GD+++ D+TY N PL +FSG NHH +++IFG+A++ DET ++Y WL + F+EA
Sbjct: 187 GDMVSLDSTYCTHSSNRPLAVFSGFNHHRKAVIFGAALLYDETTESY*WLFESFLEA 357
>CB827002
Length = 534
Score = 45.8 bits (107), Expect = 2e-05
Identities = 23/71 (32%), Positives = 38/71 (53%)
Frame = +1
Query: 358 EENPWMLEMYQKRKMWAAAHFRGKFFAGFRTTSRCEGLHSEFGKYVSALTNLHDFFQQFF 417
+++ W L +Y + A H R FFA T R + ++S F YV+A TNL+ FF+ +
Sbjct: 28 QDHEW-LSLYSSCRQGAPVHLRDTFFAEMSITQRSDSMNSYFDGYVNASTNLNQFFKLYE 204
Query: 418 RWLNYMRYREI 428
+ L +E+
Sbjct: 205 KALESRNEKEV 237
>AV409041
Length = 349
Score = 38.5 bits (88), Expect = 0.002
Identities = 26/100 (26%), Positives = 42/100 (42%), Gaps = 1/100 (1%)
Frame = +3
Query: 5 FPDREVAFMFYNWYGCFHGFAARKSRLIRN-INGEVVQQTFLCHREGIREEKYINSTSRK 63
F E A+ FY Y GF K R+ + E + F C R G +++ +
Sbjct: 51 FESHEAAYAFYKEYAKSAGFGTAKLSSRRSRASKEFIDAKFSCIRYGNKQQ-----SDDA 215
Query: 64 REHKPLSRCGCQAKVRVHIDVSSQRWYIKLFDDDHNHSFV 103
+P + GC+A + V +WY+ F +HNH +
Sbjct: 216 INPRPSPKIGCKASMHVKRRQDG-KWYVYSFVKEHNHELL 332
>AV423237
Length = 310
Score = 34.7 bits (78), Expect = 0.036
Identities = 30/99 (30%), Positives = 47/99 (47%), Gaps = 5/99 (5%)
Frame = +2
Query: 210 DGVSCMDYSIFGDVLAFDATYKKIKYNTPLVIFSGVNHHNQSIIFGSAIIGDETEDTYVW 269
D S M+YS FGD + FD T + + G + +I+ A+I +E+E ++VW
Sbjct: 17 DATSRMNYSYFGDAVIFDTTID--NHIESICSSWG*SSWATCVIWLVALIANESESSFVW 190
Query: 270 LLKIFVEAMGGKLP-----VSVITDGDLSMRNAIRKVFP 303
L + G +P S+ TD D S + + VFP
Sbjct: 191 LSGL------GSMPCLDATCSITTDLDHSYK-LLWLVFP 286
>AV769392
Length = 621
Score = 29.3 bits (64), Expect = 1.5
Identities = 29/95 (30%), Positives = 40/95 (41%), Gaps = 5/95 (5%)
Frame = +3
Query: 40 VQQTFLCHREGIREEKYINSTSRKREH--KPLSRCGCQAKVRVHI-DVSSQRWYIKLFDD 96
V + F C R I + N+TSR R H PL+ AK+ V + + +Q W
Sbjct: 189 VNRAFPCWRSSITSASHKNTTSRSRFHPPHPLNLFRSSAKL*VALRSLKNQFW------- 347
Query: 97 DHNHSFVKEKFERM--LPAHRKMSEYDKYQMNTMR 129
+K RM L A K+ D + MNT R
Sbjct: 348 ----RSLKLSLSRMCDLVAMLKLGRGDLWMMNTFR 440
>TC17583 weakly similar to UP|Q9LIE5 (Q9LIE5) Far-red impaired response
protein, mutator-like transposase-like protein,
phytochrome A signaling protein-like, partial (3%)
Length = 666
Score = 29.3 bits (64), Expect = 1.5
Identities = 12/27 (44%), Positives = 15/27 (55%)
Frame = +3
Query: 215 MDYSIFGDVLAFDATYKKIKYNTPLVI 241
MDY FGDV++ D TY PL +
Sbjct: 12 MDYEYFGDVVSLDTTYSTNNAYRPLAV 92
>TC11106
Length = 655
Score = 28.9 bits (63), Expect = 2.0
Identities = 18/51 (35%), Positives = 23/51 (44%), Gaps = 2/51 (3%)
Frame = +3
Query: 316 RNATSNIKNLHFVSKFKDCLLGDVDVDV--FQRKWEELVTEFGLEENPWML 364
R N+K+L F KFK + V V + F+ W L GL N W L
Sbjct: 105 RRTARNLKSLTFSPKFKPTIASSVPVPLVPFKECWISLFCCEGLLGNSWTL 257
>BP085116
Length = 486
Score = 27.7 bits (60), Expect = 4.4
Identities = 11/22 (50%), Positives = 13/22 (59%)
Frame = +3
Query: 192 FWRHTVHADGSLQHLFWCDGVS 213
F RHT DGS+ +FW G S
Sbjct: 72 FRRHTKRRDGSINPIFWLSGES 137
>BP029988
Length = 443
Score = 27.7 bits (60), Expect = 4.4
Identities = 11/30 (36%), Positives = 18/30 (59%)
Frame = +1
Query: 216 DYSIFGDVLAFDATYKKIKYNTPLVIFSGV 245
D+S+ + A T KK+KYN +V+ G+
Sbjct: 118 DFSVIPEXWALHXTQKKLKYNKKMVLVFGL 207
>TC10711
Length = 620
Score = 27.3 bits (59), Expect = 5.7
Identities = 17/48 (35%), Positives = 20/48 (41%)
Frame = +3
Query: 251 SIIFGSAIIGDETEDTYVWLLKIFVEAMGGKLPVSVITDGDLSMRNAI 298
S G IGD ED Y WL +I T GDL MR+ +
Sbjct: 48 STFSGKGSIGDGEEDEYAWLAEI-------------ETPGDLYMRSGV 152
>TC9631 similar to GB|AAC19270.1|3193286|T14P8 coded for by A. thaliana
cDNA H76583 {Arabidopsis thaliana;}, partial (30%)
Length = 624
Score = 27.3 bits (59), Expect = 5.7
Identities = 21/94 (22%), Positives = 36/94 (37%), Gaps = 9/94 (9%)
Frame = +2
Query: 16 NWYGCFHGFAARKSRLIRNINGEVVQQTFLCHREGIREEKYINSTSRKREHKPLSRCGCQ 75
+W GC H RK L R + + + R + IN +R++ P + C C+
Sbjct: 26 DWPGCVHSEEKRKYMLFRGV---------FKNFKRTRVWRTINDGNRRKLDLPCAFCACK 178
Query: 76 AKVRVHIDVSSQRWY---------IKLFDDDHNH 100
+H +R + ++ F DH H
Sbjct: 179 HTWDLHSAFCLRRGFGFHDDGEPVVRAFVCDHGH 280
>TC8758 similar to GB|AAC77862.2|20197419|AC005623 Argonaute (AGO1)-like
protein {Arabidopsis thaliana;} , partial (32%)
Length = 1142
Score = 26.9 bits (58), Expect = 7.4
Identities = 15/41 (36%), Positives = 21/41 (50%)
Frame = -1
Query: 281 KLPVSVITDGDLSMRNAIRKVFPEAHHRLCAWHLIRNATSN 321
KL S T+G S+ I KVFP +C HL+ + + N
Sbjct: 497 KLVPSKRTNGKQSLLPIIGKVFPIQLRTICLLHLVNHFSPN 375
>TC11996 similar to GB|AAO63323.1|28950799|BT005259 At2g03430 {Arabidopsis
thaliana;}, partial (27%)
Length = 550
Score = 26.9 bits (58), Expect = 7.4
Identities = 13/27 (48%), Positives = 15/27 (55%)
Frame = +3
Query: 62 RKREHKPLSRCGCQAKVRVHIDVSSQR 88
R +K SRCGC + RVH SS R
Sbjct: 69 RPSSYKTWSRCGCGGQGRVHSARSSYR 149
>TC20070 similar to PIR|JQ1073|JQ1073 tryptophan synthase beta-2 chain
precursor - Arabidopsis thaliana
{Arabidopsis thaliana;} , partial (14%)
Length = 453
Score = 26.6 bits (57), Expect = 9.7
Identities = 10/26 (38%), Positives = 16/26 (61%)
Frame = -3
Query: 335 LLGDVDVDVFQRKWEELVTEFGLEEN 360
+LG ++ F+ KWE+ FG+E N
Sbjct: 157 VLGAANLSRFEEKWEDGYGSFGMESN 80
>TC17208 similar to UP|Q9MAV5 (Q9MAV5) F24O1.4, partial (13%)
Length = 682
Score = 26.6 bits (57), Expect = 9.7
Identities = 18/52 (34%), Positives = 22/52 (41%)
Frame = -3
Query: 313 HLIRNATSNIKNLHFVSKFKDCLLGDVDVDVFQRKWEELVTEFGLEENPWML 364
HL+ S N H K CLLG VF R W + T G N W++
Sbjct: 431 HLVIYINSKAFNAH---KELLCLLGK---GVFFRDWRFVWTSIGSSSNSWLI 294
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.327 0.139 0.452
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,254,115
Number of Sequences: 28460
Number of extensions: 127482
Number of successful extensions: 814
Number of sequences better than 10.0: 40
Number of HSP's better than 10.0 without gapping: 804
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 810
length of query: 485
length of database: 4,897,600
effective HSP length: 94
effective length of query: 391
effective length of database: 2,222,360
effective search space: 868942760
effective search space used: 868942760
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 57 (26.6 bits)
Medicago: description of AC146755.5


