

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146755.12 + phase: 0
(57 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
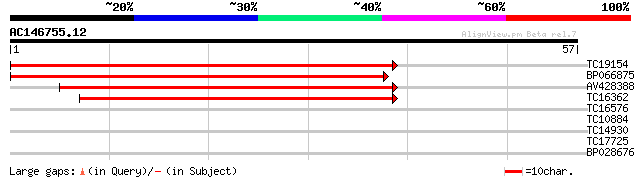
TC19154 similar to UP|AAS38575 (AAS38575) Short-chain dehydrogen... 65 3e-12
BP066875 62 3e-11
AV428388 44 5e-06
TC16362 similar to UP|HCD2_HUMAN (Q99714) 3-hydroxyacyl-CoA dehy... 41 4e-05
TC16576 weakly similar to UP|O76728 (O76728) Oligopeptidase B, p... 26 1.8
TC10884 similar to UP|Q80KM7 (Q80KM7) Envelope glycoprotein gpUL... 25 3.1
TC14930 25 4.0
TC17725 homologue to UP|Q8LLR2 (Q8LLR2) MADS-box protein 2, part... 24 6.9
BP028676 23 9.0
>TC19154 similar to UP|AAS38575 (AAS38575) Short-chain dehydrogenase Tic32,
partial (44%)
Length = 508
Score = 65.1 bits (157), Expect = 3e-12
Identities = 30/39 (76%), Positives = 34/39 (86%)
Frame = +2
Query: 1 MWPFSKKGVSGFSGNSTAEKVTHGIDASGLTAIVTGLSS 39
MWPFSKKG SGFS +STAE+VT GID +GLTA+VTG SS
Sbjct: 80 MWPFSKKGPSGFSSSSTAEQVTQGIDGTGLTAVVTGASS 196
>BP066875
Length = 521
Score = 61.6 bits (148), Expect = 3e-11
Identities = 28/38 (73%), Positives = 31/38 (80%)
Frame = +3
Query: 1 MWPFSKKGVSGFSGNSTAEKVTHGIDASGLTAIVTGLS 38
MWPF +KG SGFS +STAE VTHGID +GL AIVTG S
Sbjct: 393 MWPFCRKGASGFSSSSTAEDVTHGIDGNGLVAIVTGRS 506
>AV428388
Length = 369
Score = 44.3 bits (103), Expect = 5e-06
Identities = 22/34 (64%), Positives = 26/34 (75%)
Frame = +2
Query: 6 KKGVSGFSGNSTAEKVTHGIDASGLTAIVTGLSS 39
+ G SGF STAE+VT GIDAS LTAI+TG +S
Sbjct: 38 RHGPSGFGSASTAEQVTEGIDASNLTAIITGGAS 139
>TC16362 similar to UP|HCD2_HUMAN (Q99714) 3-hydroxyacyl-CoA dehydrogenase
type II (Type II HADH) (Endoplasmic
reticulum-associated amyloid beta-peptide binding
protein) (Short-chain type dehydrogenase/reductase
XH98G2) , partial (7%)
Length = 613
Score = 41.2 bits (95), Expect = 4e-05
Identities = 20/32 (62%), Positives = 25/32 (77%)
Frame = +2
Query: 8 GVSGFSGNSTAEKVTHGIDASGLTAIVTGLSS 39
G SGF +TAE+V+ GIDAS LTAI+TG +S
Sbjct: 119 GPSGFGSATTAEQVSEGIDASNLTAIITGGAS 214
>TC16576 weakly similar to UP|O76728 (O76728) Oligopeptidase B, partial
(30%)
Length = 1202
Score = 25.8 bits (55), Expect = 1.8
Identities = 12/52 (23%), Positives = 26/52 (49%)
Frame = -2
Query: 5 SKKGVSGFSGNSTAEKVTHGIDASGLTAIVTGLSSITLSYHFFNFKIFKHAL 56
S KG + + + ++ S A++ + S++L ++FF+ K HA+
Sbjct: 415 STKGTPATAALNKSGRILRTAPISKPPALLPSMQSLSLEHNFFSIKYSAHAI 260
>TC10884 similar to UP|Q80KM7 (Q80KM7) Envelope glycoprotein gpUL73, partial
(12%)
Length = 523
Score = 25.0 bits (53), Expect = 3.1
Identities = 8/18 (44%), Positives = 12/18 (66%)
Frame = +3
Query: 40 ITLSYHFFNFKIFKHALC 57
+ SY +F+ K+ KH LC
Sbjct: 348 VATSYDWFSLKLIKHTLC 401
>TC14930
Length = 331
Score = 24.6 bits (52), Expect = 4.0
Identities = 14/46 (30%), Positives = 22/46 (47%), Gaps = 2/46 (4%)
Frame = -3
Query: 13 SGNSTAEKVTHGIDASGLTAIVTGLSSIT--LSYHFFNFKIFKHAL 56
SGN T G ASG+ ++ + S+ LSY+ IF ++
Sbjct: 242 SGNITIRSSVEGFSASGVGSVPIKIKSLNPHLSYYSHTQSIFSFSI 105
>TC17725 homologue to UP|Q8LLR2 (Q8LLR2) MADS-box protein 2, partial (45%)
Length = 970
Score = 23.9 bits (50), Expect = 6.9
Identities = 8/19 (42%), Positives = 13/19 (68%)
Frame = +1
Query: 39 SITLSYHFFNFKIFKHALC 57
S++ ++ FFNFK+ LC
Sbjct: 31 SLSRAFKFFNFKVLNFFLC 87
>BP028676
Length = 520
Score = 23.5 bits (49), Expect = 9.0
Identities = 11/38 (28%), Positives = 17/38 (43%)
Frame = -2
Query: 7 KGVSGFSGNSTAEKVTHGIDASGLTAIVTGLSSITLSY 44
K S N T + HG+ ASG + + +IT +
Sbjct: 465 KDTSELKMNGTKSMIGHGLGASGGLEAIATIKAITTGW 352
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.322 0.135 0.415
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 965,009
Number of Sequences: 28460
Number of extensions: 8201
Number of successful extensions: 59
Number of sequences better than 10.0: 19
Number of HSP's better than 10.0 without gapping: 59
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 59
length of query: 57
length of database: 4,897,600
effective HSP length: 33
effective length of query: 24
effective length of database: 3,958,420
effective search space: 95002080
effective search space used: 95002080
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 49 (23.5 bits)
Medicago: description of AC146755.12


