

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
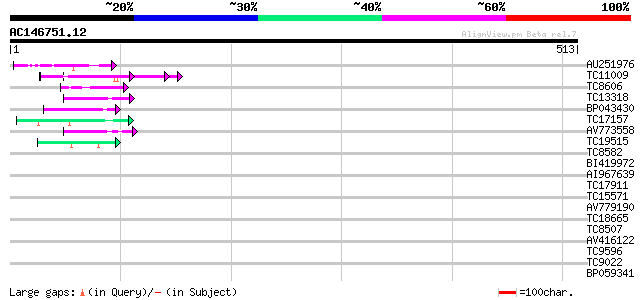
Query= AC146751.12 - phase: 0
(513 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
AU251976 63 1e-10
TC11009 similar to GB|AAP21377.1|30102918|BT006569 At1g47970 {Ar... 50 9e-07
TC8606 similar to UP|AAQ82688 (AAQ82688) Epa4p, partial (7%) 45 2e-05
TC13318 similar to UP|CAA20314 (CAA20314) SPBC30B4.01c protein (... 45 2e-05
BP043430 41 4e-04
TC17157 similar to UP|CAD27462 (CAD27462) Nucleosome assembly pr... 41 4e-04
AV773558 40 7e-04
TC19515 similar to UP|Q9YGK0 (Q9YGK0) Vitellogenin precursor, pa... 40 7e-04
TC8582 similar to UP|Q8LJS2 (Q8LJS2) Nucleolar histone deacetyla... 40 0.001
BI419972 39 0.003
AI967639 38 0.003
TC17911 weakly similar to UP|Q9M651 (Q9M651) RAN GTPase activati... 38 0.003
TC15571 weakly similar to PIR|S19652|S19652 cellodextrinase C - ... 37 0.006
AV779190 37 0.006
TC18665 similar to UP|O23061 (O23061) BAC IG005I10, partial (65%) 36 0.017
TC8507 similar to UP|Q8LJS2 (Q8LJS2) Nucleolar histone deacetyla... 36 0.017
AV416122 35 0.029
TC9596 similar to UP|AAR24722 (AAR24722) At5g49400, partial (66%) 35 0.038
TC9022 similar to PIR|T46225|T46225 alpha NAC-like protein - Ara... 34 0.050
BP059341 33 0.11
>AU251976
Length = 298
Score = 63.2 bits (152), Expect = 1e-10
Identities = 47/97 (48%), Positives = 55/97 (56%), Gaps = 4/97 (4%)
Frame = +2
Query: 4 ATKKKPSKPVKPQKAPKKVQIVPIKDDSTSESEEEEAPPLQHLELDGSDSDLS----SDG 59
ATK++ P+KP+K PK P K+DS +ESEEE AP L L D D S SDG
Sbjct: 47 ATKQQRKSPMKPKK-PK--HFTP-KEDSPAESEEE-APQLLELGSDDGGGDDSASDFSDG 211
Query: 60 DDQLADDFLQGSDDEDNDNDNDDEGSDLESGSGSDED 96
DD L DDFLQGSD D+EG S SGS+ D
Sbjct: 212 DDPLVDDFLQGSD--------DNEGKASASASGSESD 298
>TC11009 similar to GB|AAP21377.1|30102918|BT006569 At1g47970 {Arabidopsis
thaliana;}, partial (44%)
Length = 700
Score = 50.1 bits (118), Expect = 9e-07
Identities = 31/100 (31%), Positives = 48/100 (48%), Gaps = 4/100 (4%)
Frame = +2
Query: 49 DGSDSDLSSDGDDQLADDFLQGSDDEDNDNDNDDEGSDLESGSGS----DEDEDSDSESG 104
D D DGD+ DD G DD+D++ D ++EG +E G G D+D+D D +
Sbjct: 32 DDDDDGEDDDGDEDDDDDAPGGGDDDDDEEDEEEEGG-VEGGRGGGGDPDDDDDDDDDDD 208
Query: 105 EEDIARKSKLIDRKRERDSQAAADELQTNIQTNIQDESDE 144
EE+ + + R AA DE ++ +DE D+
Sbjct: 209 EEEEEEEDLGTEYLVRRTVAAAEDEEASSDFEPEEDEGDD 328
Score = 41.2 bits (95), Expect = 4e-04
Identities = 27/129 (20%), Positives = 61/129 (46%)
Frame = +2
Query: 28 KDDSTSESEEEEAPPLQHLELDGSDSDLSSDGDDQLADDFLQGSDDEDNDNDNDDEGSDL 87
+DD E ++++AP G D D D +++ + +G + +D+D+DD+
Sbjct: 50 EDDDGDEDDDDDAPG------GGDDDDDEEDEEEEGGVEGGRGGGGDPDDDDDDDD---- 199
Query: 88 ESGSGSDEDEDSDSESGEEDIARKSKLIDRKRERDSQAAADELQTNIQTNIQDESDEFTL 147
DE+E+ + + G E + R++ E S +E + + N D+ ++ +
Sbjct: 200 ----DDDEEEEEEEDLGTEYLVRRTVAAAEDEEASSDFEPEEDEGD--DNDNDDGEKAGV 361
Query: 148 PTKQELEEE 156
P+K++ ++
Sbjct: 362 PSKRKRSDK 388
Score = 41.2 bits (95), Expect = 4e-04
Identities = 29/88 (32%), Positives = 44/88 (49%), Gaps = 3/88 (3%)
Frame = +2
Query: 29 DDSTSESEEEEAPPLQHLELDGSDSDLSSDGDDQLADDFLQGSDD-EDNDNDNDDEGSDL 87
DD E EEEE ++L +++ D++ + DF D+ +DNDND+ ++
Sbjct: 197 DDDDEEEEEEEDLGTEYLVR----RTVAAAEDEEASSDFEPEEDEGDDNDNDDGEKAGVP 364
Query: 88 ESGSGSDED--EDSDSESGEEDIARKSK 113
SD+D D DS+ G ED R SK
Sbjct: 365 SKRKRSDKDGSGDDDSDDGGEDDERPSK 448
Score = 36.2 bits (82), Expect = 0.013
Identities = 21/56 (37%), Positives = 28/56 (49%)
Frame = +2
Query: 51 SDSDLSSDGDDQLADDFLQGSDDEDNDNDNDDEGSDLESGSGSDEDEDSDSESGEE 106
S + +S D DD DD D ED+D D DD+ G D++ED + E G E
Sbjct: 5 SRTRMSGDEDD---DD-----DGEDDDGDEDDDDDAPGGGDDDDDEEDEEEEGGVE 148
>TC8606 similar to UP|AAQ82688 (AAQ82688) Epa4p, partial (7%)
Length = 1008
Score = 45.4 bits (106), Expect = 2e-05
Identities = 23/61 (37%), Positives = 35/61 (56%)
Frame = -2
Query: 47 ELDGSDSDLSSDGDDQLADDFLQGSDDEDNDNDNDDEGSDLESGSGSDEDEDSDSESGEE 106
E +G D + DG+DQ DD D+D D+D +D+G + E G +E+++ D E EE
Sbjct: 443 EENGEDEE-DEDGEDQEDDD------DDDEDDDEEDDGGEDEEEEGVEEEDNEDEEEDEE 285
Query: 107 D 107
D
Sbjct: 284 D 282
Score = 40.0 bits (92), Expect = 0.001
Identities = 34/120 (28%), Positives = 57/120 (47%), Gaps = 11/120 (9%)
Frame = -2
Query: 29 DDSTSESEEEEAPPLQHLELDGSDSDLSSDGDDQLADDFLQGSDDEDNDNDNDDEGSDLE 88
++S ++S ++AP G+ + +G+D+ +D G D ED+D+D DE D E
Sbjct: 512 NNSNNKSNSKKAPEGG----PGAGAGPEENGEDEEDED---GEDQEDDDDD--DEDDDEE 360
Query: 89 SGSGSDED----EDSDSESGEEDIARKSKLIDRKRERDSQAAADE-------LQTNIQTN 137
G DE+ E+ D+E EED + L K+ + A+ L TN+ +N
Sbjct: 359 DDGGEDEEEEGVEEEDNEDEEEDEEDEEALQPPKKRKK*TTLAESSLLLFVLLCTNLDSN 180
Score = 31.6 bits (70), Expect = 0.32
Identities = 29/139 (20%), Positives = 54/139 (37%), Gaps = 6/139 (4%)
Frame = -2
Query: 29 DDSTSESEEEEAP------PLQHLELDGSDSDLSSDGDDQLADDFLQGSDDEDNDNDNDD 82
DD E ++ + P L + G ++ ++ + + A + G+ +N D+
Sbjct: 602 DDEGDEDDDGDGPFGEGEEDLSSEDGAGYGNNSNNKSNSKKAPEGGPGAGAGPEENGEDE 423
Query: 83 EGSDLESGSGSDEDEDSDSESGEEDIARKSKLIDRKRERDSQAAADELQTNIQTNIQDES 142
E D E D+D++ D E + DE + ++ +D
Sbjct: 422 EDEDGEDQEDDDDDDEDDDE-------------------EDDGGEDEEEEGVEE--EDNE 306
Query: 143 DEFTLPTKQELEEEALRPP 161
DE + E +EEAL+PP
Sbjct: 305 DE----EEDEEDEEALQPP 261
>TC13318 similar to UP|CAA20314 (CAA20314) SPBC30B4.01c protein (SPBC3D6.14C
protein) (Fragment), partial (15%)
Length = 543
Score = 45.4 bits (106), Expect = 2e-05
Identities = 20/65 (30%), Positives = 37/65 (56%)
Frame = +3
Query: 49 DGSDSDLSSDGDDQLADDFLQGSDDEDNDNDNDDEGSDLESGSGSDEDEDSDSESGEEDI 108
DG + +D +++ DD + +DED+D+D+DD+ E D+D+D + E G E++
Sbjct: 219 DGPPKPVQTDNEEEEDDDEEEDDEDEDDDDDDDDDDDGEEE---EDDDDDDEEEEGSEEV 389
Query: 109 ARKSK 113
+ K
Sbjct: 390 EVEGK 404
Score = 36.6 bits (83), Expect = 0.010
Identities = 25/92 (27%), Positives = 38/92 (41%)
Frame = +3
Query: 13 VKPQKAPKKVQIVPIKDDSTSESEEEEAPPLQHLELDGSDSDLSSDGDDQLADDFLQGSD 72
+ P PK P++ D+ E +++E E D D D D DD D
Sbjct: 210 IYPDGPPK-----PVQTDNEEEEDDDE-------EEDDEDEDDDDDDDD----------D 323
Query: 73 DEDNDNDNDDEGSDLESGSGSDEDEDSDSESG 104
D+ + ++DD+ + E GS E E D G
Sbjct: 324 DDGEEEEDDDDDDEEEEGSEEVEVEGKDGAKG 419
Score = 34.3 bits (77), Expect = 0.050
Identities = 16/56 (28%), Positives = 28/56 (49%)
Frame = +3
Query: 52 DSDLSSDGDDQLADDFLQGSDDEDNDNDNDDEGSDLESGSGSDEDEDSDSESGEED 107
D ++ DG + + +D+D + D++DE D D+D+D D E E+D
Sbjct: 201 DPEIYPDGPPKPVQTDNEEEEDDDEEEDDEDEDDD------DDDDDDDDGEEEEDD 350
Score = 28.5 bits (62), Expect = 2.7
Identities = 13/46 (28%), Positives = 22/46 (47%)
Frame = +3
Query: 79 DNDDEGSDLESGSGSDEDEDSDSESGEEDIARKSKLIDRKRERDSQ 124
DN++E D E DED+D D + ++ + D + E S+
Sbjct: 246 DNEEEEDDDEEEDDEDEDDDDDDDDDDDGEEEEDDDDDDEEEEGSE 383
>BP043430
Length = 493
Score = 41.2 bits (95), Expect = 4e-04
Identities = 24/71 (33%), Positives = 42/71 (58%), Gaps = 1/71 (1%)
Frame = -1
Query: 31 STSESEEEEAPPLQHLELDGSDSDLSS-DGDDQLADDFLQGSDDEDNDNDNDDEGSDLES 89
+TS + P ++ G D++ S D DD+ D+ + D+E+ D+++DD+GS ES
Sbjct: 439 TTSYAAPPPPKPKIPVKASGDDNEGSGKDNDDEGNDNEVDEEDEEEEDSNDDDDGS--ES 266
Query: 90 GSGSDEDEDSD 100
+D++EDSD
Sbjct: 265 EEFNDDEEDSD 233
Score = 38.1 bits (87), Expect = 0.003
Identities = 18/38 (47%), Positives = 24/38 (62%), Gaps = 1/38 (2%)
Frame = -1
Query: 70 GSDDEDNDNDNDDEGSDLESGSGSDEDEDS-DSESGEE 106
G D+E + DNDDEG+D E +E+EDS D + G E
Sbjct: 382 GDDNEGSGKDNDDEGNDNEVDEEDEEEEDSNDDDDGSE 269
Score = 37.0 bits (84), Expect = 0.008
Identities = 22/67 (32%), Positives = 32/67 (46%)
Frame = -1
Query: 41 PPLQHLELDGSDSDLSSDGDDQLADDFLQGSDDEDNDNDNDDEGSDLESGSGSDEDEDSD 100
PP + + S D G D +DDE NDN+ D+E D E +D+D+ S+
Sbjct: 415 PPKPKIPVKASGDDNEGSGKD---------NDDEGNDNEVDEE--DEEEEDSNDDDDGSE 269
Query: 101 SESGEED 107
SE +D
Sbjct: 268 SEEFNDD 248
>TC17157 similar to UP|CAD27462 (CAD27462) Nucleosome assembly protein
1-like protein 3, partial (81%)
Length = 1298
Score = 41.2 bits (95), Expect = 4e-04
Identities = 36/130 (27%), Positives = 50/130 (37%), Gaps = 24/130 (18%)
Frame = +2
Query: 7 KKPSKPVKPQKAPKKVQI---------VPIKDDSTSESEEEEAPPLQHLELD-GSD---- 52
KK SK KP +K + VP DD + EE L + D GS
Sbjct: 530 KKGSKNTKPITKTEKCESFFNFFNPPQVPEDDDDIDDDAVEELQNLMEHDYDIGSTIRDK 709
Query: 53 ----------SDLSSDGDDQLADDFLQGSDDEDNDNDNDDEGSDLESGSGSDEDEDSDSE 102
+ + + +D G +DED D+D+D+E D D+D+D D E
Sbjct: 710 IIPHAVSWFTGEAEQSDFEDIEEDDEDGDEDEDEDDDDDEEEED-------DDDDDEDDE 868
Query: 103 SGEEDIARKS 112
GE KS
Sbjct: 869 EGEGKSKSKS 898
Score = 37.0 bits (84), Expect = 0.008
Identities = 26/88 (29%), Positives = 37/88 (41%)
Frame = +2
Query: 37 EEEAPPLQHLELDGSDSDLSSDGDDQLADDFLQGSDDEDNDNDNDDEGSDLESGSGSDED 96
E E + +E D D D D DD DD++ + D+DD+ D E G G
Sbjct: 743 EAEQSDFEDIEEDDEDGDEDEDEDD----------DDDEEEEDDDDDDEDDEEGEGK--- 883
Query: 97 EDSDSESGEEDIARKSKLIDRKRERDSQ 124
S S+SG + K + +R E Q
Sbjct: 884 --SKSKSGSKARPGKDQPTERPPECKQQ 961
>AV773558
Length = 469
Score = 40.4 bits (93), Expect = 7e-04
Identities = 26/68 (38%), Positives = 39/68 (57%), Gaps = 1/68 (1%)
Frame = -2
Query: 49 DGSDSDLSSDGDDQLADDF-LQGSDDEDNDNDNDDEGSDLESGSGSDEDEDSDSESGEED 107
D D ++GD++L DDF L S+D+D+D+D+DD+ D DEDE+ E G
Sbjct: 438 DSPDGLFFNEGDEELDDDFVLVYSEDDDDDDDDDDDDDDYT----DDEDEE---EGGFYS 280
Query: 108 IARKSKLI 115
+ K +LI
Sbjct: 279 L*MKLELI 256
>TC19515 similar to UP|Q9YGK0 (Q9YGK0) Vitellogenin precursor, partial (3%)
Length = 504
Score = 40.4 bits (93), Expect = 7e-04
Identities = 31/106 (29%), Positives = 39/106 (36%), Gaps = 31/106 (29%)
Frame = +3
Query: 26 PIKDDSTSESEEEEAPPLQHLELDGSDSD--------LSSDGDDQLADDFLQGSDDEDND 77
P DDS + E E + DG D D SD DD DD+ SD +D D
Sbjct: 51 PTDDDSDPDDAESEDEEDDDDDDDGDDEDPLLGPGLGRESDSDDMSHDDYDSASDIDDED 230
Query: 78 ND-----------------------NDDEGSDLESGSGSDEDEDSD 100
D +DD+ +LES S DED D +
Sbjct: 231 ADFILDDLDFDGGTGLLEIVGDRDEDDDDSQELESASSDDEDFDDN 368
Score = 36.6 bits (83), Expect = 0.010
Identities = 31/112 (27%), Positives = 48/112 (42%), Gaps = 6/112 (5%)
Frame = +3
Query: 60 DDQLADDFLQGSDDEDNDNDNDDEGSDLESGSGSDEDEDSDSESGEEDIARKSKLIDRKR 119
DD D + D+ED+D+D+D + D G G + DSD S +D S + D
Sbjct: 57 DDDSDPDDAESEDEEDDDDDDDGDDEDPLLGPGLGRESDSDDMS-HDDYDSASDIDD--- 224
Query: 120 ERDSQAAADELQTNIQTNI------QDESDEFTLPTKQELEEEALRPPDLSN 165
D+ D+L + T + +DE D+ QELE + D +
Sbjct: 225 -EDADFILDDLDFDGGTGLLEIVGDRDEDDD----DSQELESASSDDEDFDD 365
>TC8582 similar to UP|Q8LJS2 (Q8LJS2) Nucleolar histone deacetylase
HD2-P39, partial (65%)
Length = 1341
Score = 39.7 bits (91), Expect = 0.001
Identities = 38/132 (28%), Positives = 60/132 (44%), Gaps = 7/132 (5%)
Frame = +1
Query: 3 PATKKKPSKPVKPQK-------APKKVQIVPIKDDSTSESEEEEAPPLQHLELDGSDSDL 55
P TK + +K K K + K+V+IV K D EEEA +LD S D+
Sbjct: 436 PETKTEDAKVAKTAKPAAAAGPSAKQVKIVDPKKD------EEEA------DLDVSSPDV 579
Query: 56 SSDGDDQLADDFLQGSDDEDNDNDNDDEGSDLESGSGSDEDEDSDSESGEEDIARKSKLI 115
S DD + S DED+D+++DDE D EE+ +K +
Sbjct: 580 SGYEDDLI-------SADEDSDDESDDESDD------------------EEETPTPAKKV 684
Query: 116 DRKRERDSQAAA 127
D+ ++R +++A+
Sbjct: 685 DQGKKRPNESAS 720
>BI419972
Length = 498
Score = 38.5 bits (88), Expect = 0.003
Identities = 16/44 (36%), Positives = 24/44 (54%)
Frame = +1
Query: 60 DDQLADDFLQGSDDEDNDNDNDDEGSDLESGSGSDEDEDSDSES 103
D + DD +D+D D+D+DDEG D G + D + D E+
Sbjct: 343 DSETEDDEDGDDEDDDADDDDDDEGEDYSGDEGEEADPEDDPEA 474
Score = 32.0 bits (71), Expect = 0.25
Identities = 15/57 (26%), Positives = 32/57 (55%)
Frame = +1
Query: 51 SDSDLSSDGDDQLADDFLQGSDDEDNDNDNDDEGSDLESGSGSDEDEDSDSESGEED 107
S+++ DGDD+ DD DD++ ++ + DEG + + ++D +++ G +D
Sbjct: 346 SETEDDEDGDDE--DDDADDDDDDEGEDYSGDEGEEAD----PEDDPEANGAGGSDD 498
Score = 31.2 bits (69), Expect = 0.42
Identities = 9/34 (26%), Positives = 24/34 (70%)
Frame = +1
Query: 73 DEDNDNDNDDEGSDLESGSGSDEDEDSDSESGEE 106
++D++ ++D++G D + + D+D++ + SG+E
Sbjct: 337 NKDSETEDDEDGDDEDDDADDDDDDEGEDYSGDE 438
>AI967639
Length = 335
Score = 38.1 bits (87), Expect = 0.003
Identities = 24/69 (34%), Positives = 36/69 (51%), Gaps = 5/69 (7%)
Frame = +1
Query: 72 DDEDNDNDNDDEGSDLESGSGSDEDEDSD----SESGEEDIARKSK-LIDRKRERDSQAA 126
D+ED + D DE +LE D+D+D D SE + D RK K L R R + +
Sbjct: 103 DEEDEEEDQGDE-YNLEDEEEDDDDDDDDDLSISEDSDSDKPRKVKQLPGRTRRETKRRS 279
Query: 127 ADELQTNIQ 135
DE+Q+ ++
Sbjct: 280 VDEIQSGLR 306
>TC17911 weakly similar to UP|Q9M651 (Q9M651) RAN GTPase activating protein
2, partial (10%)
Length = 531
Score = 38.1 bits (87), Expect = 0.003
Identities = 21/59 (35%), Positives = 30/59 (50%)
Frame = +2
Query: 55 LSSDGDDQLADDFLQGSDDEDNDNDNDDEGSDLESGSGSDEDEDSDSESGEEDIARKSK 113
+S++G D+L D D NDND +G D DE+SD E GE+++ K K
Sbjct: 89 ISNEGIDELTDILKNSPDVLGPLNDNDPDGED--------NDEESDEEDGEDELESKMK 241
>TC15571 weakly similar to PIR|S19652|S19652 cellodextrinase C - Pseudomonas
fluorescens {Pseudomonas fluorescens;} , partial (6%)
Length = 784
Score = 37.4 bits (85), Expect = 0.006
Identities = 25/109 (22%), Positives = 51/109 (45%)
Frame = -2
Query: 71 SDDEDNDNDNDDEGSDLESGSGSDEDEDSDSESGEEDIARKSKLIDRKRERDSQAAADEL 130
+D +D D+D+D+E +D G +E+ DS +E+IA D+ A+A+
Sbjct: 771 TDSDDQDDDDDEEETD-SFDQGDEEETDSSLSHSDEEIAL----------LDTHASAE-- 631
Query: 131 QTNIQTNIQDESDEFTLPTKQELEEEALRPPDLSNLQRRIKEIVRVLSN 179
+P Q EE+ + PP +++ + ++E+ ++ +N
Sbjct: 630 ---------------GVPVAQPAEEQPIPPPPTTSMSQLLQELQKLRAN 529
>AV779190
Length = 539
Score = 37.4 bits (85), Expect = 0.006
Identities = 34/119 (28%), Positives = 55/119 (45%), Gaps = 9/119 (7%)
Frame = +3
Query: 5 TKKKPSKPVKPQKAPKKVQIV-----PIKDDSTSESE----EEEAPPLQHLELDGSDSDL 55
T++K + P P K V + P DS +S+ EEE + + D
Sbjct: 147 TQRKKTGPRLPSS*QK*VNGLNPAPEPSSVDSDDQSDFYEYEEERAEEESRKNKRYDPVF 326
Query: 56 SSDGDDQLADDFLQGSDDEDNDNDNDDEGSDLESGSGSDEDEDSDSESGEEDIARKSKL 114
+D D +D + SDDE+ D+D+D G+ + + SD+D SGEED R +++
Sbjct: 327 VNDELDSEIEDEIVPSDDENEDDDDDYFGTKRNANAQSDDD------SGEEDDGRHARM 485
>TC18665 similar to UP|O23061 (O23061) BAC IG005I10, partial (65%)
Length = 451
Score = 35.8 bits (81), Expect = 0.017
Identities = 23/61 (37%), Positives = 31/61 (50%), Gaps = 7/61 (11%)
Frame = -2
Query: 66 DFLQGSDDEDNDNDND----DEGSDLESGSGSDEDEDSDSES---GEEDIARKSKLIDRK 118
DF G DED D D+D +E D + G G +EDE ++ S G+E R+ D K
Sbjct: 219 DFESGEGDEDEDLDSDGFRDEEIGDDDEGEGEEEDEATEEASGFDGDEHEVRERAPGDEK 40
Query: 119 R 119
R
Sbjct: 39 R 37
>TC8507 similar to UP|Q8LJS2 (Q8LJS2) Nucleolar histone deacetylase
HD2-P39, partial (40%)
Length = 620
Score = 35.8 bits (81), Expect = 0.017
Identities = 24/80 (30%), Positives = 39/80 (48%)
Frame = +3
Query: 65 DDFLQGSDDEDNDNDNDDEGSDLESGSGSDEDEDSDSESGEEDIARKSKLIDRKRERDSQ 124
D+ D++D D+D+D+E D +S S ++SDS+ +E+ K +KR DS
Sbjct: 3 DEEDDSDDEDDEDDDSDEEMDDADSDS-----DESDSDDTDEETPVKKADQGKKRANDS- 164
Query: 125 AAADELQTNIQTNIQDESDE 144
A + T NI E +
Sbjct: 165 ALKTPVSTKKAKNITPEKTD 224
>AV416122
Length = 414
Score = 35.0 bits (79), Expect = 0.029
Identities = 29/114 (25%), Positives = 48/114 (41%), Gaps = 7/114 (6%)
Frame = +3
Query: 7 KKPSKPVKPQKAPKKVQIVPIKDDSTSESEEEEAPPLQHLELDGSDSDLSSDGDDQLADD 66
++P+ P K P + P K + + E P + ++ S DD +
Sbjct: 81 QEPTFPSKRHPVPDPQEQPPTKAPKLAATPE----PNPDSNDNNNNHPAVSATDDCSSQP 248
Query: 67 FLQGSDDEDNDND-------NDDEGSDLESGSGSDEDEDSDSESGEEDIARKSK 113
+DD + DND +D+ D E ++DED DS +GE ++ RK K
Sbjct: 249 KEAANDDNEPDNDVQFEGEEEEDDEEDKEEEDEEEDDEDDDS-NGEAEVDRKGK 407
>TC9596 similar to UP|AAR24722 (AAR24722) At5g49400, partial (66%)
Length = 1196
Score = 34.7 bits (78), Expect = 0.038
Identities = 27/96 (28%), Positives = 44/96 (45%), Gaps = 2/96 (2%)
Frame = +2
Query: 51 SDSDLSSDGDDQLADDFLQGSDDEDNDNDNDDEGSDLESGSGSDEDEDSDSESGEEDIAR 110
SDSD SD +D + E + GSD SGS SD S S+S EE R
Sbjct: 641 SDSDSGSDSEDSVF---------ETESGSSSVTGSDYSSGSSSD---SSSSDSEEERRRR 784
Query: 111 KSKLIDRKRE--RDSQAAADELQTNIQTNIQDESDE 144
+ K +K++ R + ++ ++ ++ +SD+
Sbjct: 785 RKKKTQKKKKGRRHKKYSSSSESSDSESASDSDSDD 892
>TC9022 similar to PIR|T46225|T46225 alpha NAC-like protein - Arabidopsis
thaliana {Arabidopsis thaliana;}, partial (75%)
Length = 1083
Score = 34.3 bits (77), Expect = 0.050
Identities = 28/104 (26%), Positives = 44/104 (41%), Gaps = 3/104 (2%)
Frame = +3
Query: 2 APATKKKPSKPVKPQKAPKKVQIVPIKDDSTSESEEEEAPPLQHLELDGSDSDLSSDG-- 59
+P K +PS V AP P+ D S+ E+ + L D
Sbjct: 57 SPRLKPQPSH-VSQNMAPG-----PVVDASSEAEALEQVLSAEETTLKKKPQPQEDDAPI 218
Query: 60 -DDQLADDFLQGSDDEDNDNDNDDEGSDLESGSGSDEDEDSDSE 102
+D DD + DDED D+D++D+ + + GS+ + S SE
Sbjct: 219 VEDVKDDDKDEADDDEDEDDDDEDDDKEDGALGGSEGSKQSRSE 350
>BP059341
Length = 471
Score = 33.1 bits (74), Expect = 0.11
Identities = 17/44 (38%), Positives = 25/44 (56%)
Frame = -3
Query: 70 GSDDEDNDNDNDDEGSDLESGSGSDEDEDSDSESGEEDIARKSK 113
G ++E+ D+D+DD+ S +D DSD + G ED R SK
Sbjct: 397 GEEEEEGDDDDDDKAEVPPKRKRSSKD-DSDDDDGGEDDERPSK 269
Score = 29.3 bits (64), Expect = 1.6
Identities = 18/57 (31%), Positives = 27/57 (46%)
Frame = -3
Query: 35 SEEEEAPPLQHLELDGSDSDLSSDGDDQLADDFLQGSDDEDNDNDNDDEGSDLESGS 91
+EEEE E +G + + D DD A+ + +D+D+DD G D E S
Sbjct: 442 AEEEEESSDFEPEENGEEEEEGDDDDDDKAEVPPKRKRSSKDDSDDDDGGEDDERPS 272
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.313 0.133 0.370
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,718,640
Number of Sequences: 28460
Number of extensions: 78166
Number of successful extensions: 1085
Number of sequences better than 10.0: 113
Number of HSP's better than 10.0 without gapping: 567
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 763
length of query: 513
length of database: 4,897,600
effective HSP length: 94
effective length of query: 419
effective length of database: 2,222,360
effective search space: 931168840
effective search space used: 931168840
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 57 (26.6 bits)
Medicago: description of AC146751.12


