

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146743.8 - phase: 0
(298 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
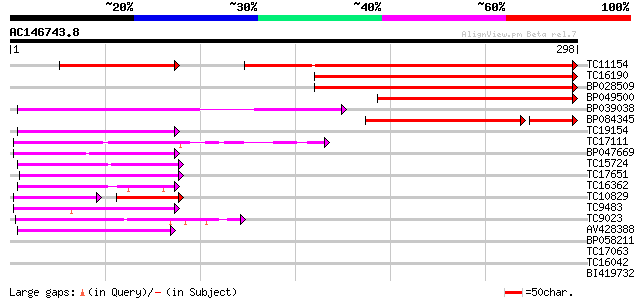
Score E
Sequences producing significant alignments: (bits) Value
TC11154 weakly similar to UP|Q9LQ75 (Q9LQ75) T1N6.22 protein, pa... 163 7e-57
TC16190 weakly similar to UP|Q9LQ75 (Q9LQ75) T1N6.22 protein, pa... 128 1e-30
BP028509 121 1e-28
BP049500 100 3e-22
BP039038 100 4e-22
BP084345 84 9e-19
TC19154 similar to UP|AAS38575 (AAS38575) Short-chain dehydrogen... 50 4e-07
TC17111 weakly similar to GB|AAL99238.1|22651517|AY082345 short-... 47 5e-06
BP047669 46 7e-06
TC15724 similar to UP|Q9LHT0 (Q9LHT0) Short chain alcohol dehydr... 45 1e-05
TC17651 weakly similar to UP|Q8KBL5 (Q8KBL5) Oxidoreductase, sho... 45 1e-05
TC16362 similar to UP|HCD2_HUMAN (Q99714) 3-hydroxyacyl-CoA dehy... 45 2e-05
TC10829 similar to UP|Q93ZA0 (Q93ZA0) AT4g13250/F17N18_140, part... 34 7e-05
TC9483 similar to UP|Q9FK50 (Q9FK50) Brn1-like protein, partial ... 41 3e-04
TC9023 weakly similar to UP|Q9FXD7 (Q9FXD7) F12A21.14, partial (... 40 6e-04
AV428388 39 8e-04
BP058211 39 0.001
TC17063 weakly similar to UP|AAS50608 (AAS50608) ABL163Wp, parti... 38 0.002
TC16042 weakly similar to UP|Q8H0D9 (Q8H0D9) Alcohol dehydroge, ... 37 0.003
BI419732 37 0.005
>TC11154 weakly similar to UP|Q9LQ75 (Q9LQ75) T1N6.22 protein, partial (68%)
Length = 833
Score = 163 bits (412), Expect(2) = 7e-57
Identities = 83/175 (47%), Positives = 113/175 (64%)
Frame = +3
Query: 124 LTQTYKKAEECLNTNYYGVKRVTMALLPLLQLSPAKARIVNLSSLRGELKRIPNERLRNE 183
+ QTY+ AEECL TNYYG K T +LLPLL LS + RIVN+SS G+L+ + E +
Sbjct: 285 IPQTYELAEECLQTNYYGAKITTESLLPLLHLSDSP-RIVNVSSSLGQLECVKKEWAKGV 461
Query: 184 LGDVDELSEGKIDAMVKKFLHDFKANDHEANGWGMMLPAYSISKASLNAYTRVLAKKNPH 243
DVD L+E +D ++K+FL DF+ E+ GW + AY +SKA++NAYTR+LA+K P
Sbjct: 462 FSDVDNLTEETVDGVLKRFLKDFQQGSLESKGWPKIFSAYIVSKAAMNAYTRILARKYPT 641
Query: 244 MLINCVHPGFVSTDFNWHKGTMTVDEGARGPVMLSLLPADGPTGCYFDCTEIAEF 298
IN V PGFV TD + G +TV+EGA PV L+L P P+G ++ E+ F
Sbjct: 642 FCINSVCPGFVKTDITANNGLLTVEEGAASPVRLALFPNGSPSGLFYHRGEVLPF 806
Score = 73.9 bits (180), Expect(2) = 7e-57
Identities = 38/64 (59%), Positives = 49/64 (76%), Gaps = 1/64 (1%)
Frame = +2
Query: 27 VTVVLTARNDTRGRDAITKLHQTGLSN-VMFHQLDVLDALSIESLAKFIQHKFGRLDILI 85
+ VVLTAR++ RG A+ L +GLS+ V+FHQLDV DA S SLA F++ KFG+LDIL+
Sbjct: 2 IKVVLTARDEKRGLQALETLKDSGLSDLVVFHQLDVADAASAASLADFVKSKFGKLDILV 181
Query: 86 NNAG 89
NNAG
Sbjct: 182NNAG 193
>TC16190 weakly similar to UP|Q9LQ75 (Q9LQ75) T1N6.22 protein, partial (41%)
Length = 1221
Score = 128 bits (321), Expect = 1e-30
Identities = 63/138 (45%), Positives = 89/138 (63%)
Frame = +1
Query: 161 RIVNLSSLRGELKRIPNERLRNELGDVDELSEGKIDAMVKKFLHDFKANDHEANGWGMML 220
RIVN+SS G+L+ +PN + L D D L+E +D ++KKFL DF+ E GW +
Sbjct: 25 RIVNVSSFLGKLECVPNGWAKGVLSDADNLTEETVDGVLKKFLQDFQDGSLEGKGWPKIF 204
Query: 221 PAYSISKASLNAYTRVLAKKNPHMLINCVHPGFVSTDFNWHKGTMTVDEGARGPVMLSLL 280
AY +SKA++NAYTR+LA+K P IN V PG+V TD + G +TV+EGA PV L+LL
Sbjct: 205 SAYIMSKAAMNAYTRILARKYPTFCINSVCPGWVKTDITANNGLLTVEEGAATPVRLALL 384
Query: 281 PADGPTGCYFDCTEIAEF 298
P P+G ++ E++ F
Sbjct: 385 PNGSPSGLFYHRGEVSSF 438
>BP028509
Length = 551
Score = 121 bits (304), Expect = 1e-28
Identities = 60/138 (43%), Positives = 88/138 (63%)
Frame = -3
Query: 161 RIVNLSSLRGELKRIPNERLRNELGDVDELSEGKIDAMVKKFLHDFKANDHEANGWGMML 220
RIVN+SS G+L+ +PN + L DVD L+E +D ++K+FL DF+ E+ GW L
Sbjct: 540 RIVNVSSSLGKLECVPNGWAKGVLSDVDNLTEEAVDGVLKRFLKDFQQGSLESKGWPKTL 361
Query: 221 PAYSISKASLNAYTRVLAKKNPHMLINCVHPGFVSTDFNWHKGTMTVDEGARGPVMLSLL 280
AY +SKA++NAYTR+LA+K P IN V G+ TD + G +T +EGA PV L+LL
Sbjct: 360 SAYIMSKAAMNAYTRILARKYPTFCINSVCXGYCKTDITVNNGFLTAEEGAASPVRLALL 181
Query: 281 PADGPTGCYFDCTEIAEF 298
P P+G ++ +++ F
Sbjct: 180 PLGSPSGLFYYKGDVSSF 127
>BP049500
Length = 518
Score = 100 bits (249), Expect = 3e-22
Identities = 51/106 (48%), Positives = 67/106 (63%), Gaps = 1/106 (0%)
Frame = -3
Query: 194 KIDAMVKKFLHDFKANDHEANGWGMMLPAYSISKASLNAYTRVLAKKNPHMLINCVHPGF 253
KID + ++L DFK EA GW ++ AY+ISK +LNAY R+LA+K P IN V PG+
Sbjct: 513 KIDEFLNEYLKDFKEGSLEAQGWPHIMSAYTISKVALNAYPRILAQKYPSFCINAVCPGY 334
Query: 254 VSTDFNWHKGTMTVDEGARGPVMLSLLP-ADGPTGCYFDCTEIAEF 298
V TD N+H G +T DEGA V L+LLP P+G +F +E F
Sbjct: 333 VQTDINYHTGLLTPDEGAEAAVRLALLPDGSSPSGPFFYRSEEKPF 196
>BP039038
Length = 538
Score = 100 bits (248), Expect = 4e-22
Identities = 69/176 (39%), Positives = 95/176 (53%), Gaps = 3/176 (1%)
Frame = +3
Query: 5 AVVTGANKGIGLEIVKQLAFLGVTVVLTARNDTRGRDAITKLHQTGLS-NVMFHQLDVLD 63
AVVTG N+GIG +VK+LA LG+ VVLTARN+ G A+ L GL+ N+ F QLDV D
Sbjct: 87 AVVTGGNRGIGFALVKRLAELGLNVVLTARNNQNGEAAMENLRAQGLAQNIHFLQLDVSD 266
Query: 64 ALSIESLAKFIQHKFG-RLDILINNAGASCVEVDKEGLKALNVDPATWLAGKVSNTLLQG 122
SI + A + +FG LDIL+NNAG S ++++ +
Sbjct: 267 PASITAFASSFKAQFGPTLDILVNNAGVSFNDLNENSV---------------------- 380
Query: 123 VLTQTYKKAEECLNTNYYGVKRVTMALLPLLQLSPA-KARIVNLSSLRGELKRIPN 177
AE + TN+YG K + ALLPL + S + RI+N+SS G L ++ N
Sbjct: 381 ------DHAESVIKTNFYGPKLLIEALLPLFRSSSSFGTRILNVSSRLGSLNKMRN 530
>BP084345
Length = 366
Score = 84.3 bits (207), Expect(2) = 9e-19
Identities = 40/84 (47%), Positives = 56/84 (66%)
Frame = -3
Query: 188 DELSEGKIDAMVKKFLHDFKANDHEANGWGMMLPAYSISKASLNAYTRVLAKKNPHMLIN 247
D L+E +D ++KKFL DF+ E+ GW + AY +SKA++NAYTR+LA+K IN
Sbjct: 364 DNLTEEAVDGVLKKFLKDFQQGSLESKGWPKVFSAYIMSKAAMNAYTRILARKYTTFCIN 185
Query: 248 CVHPGFVSTDFNWHKGTMTVDEGA 271
V PG+V TD + G +TV+EGA
Sbjct: 184 SVCPGYVKTDVTNNNGLLTVEEGA 113
Score = 25.0 bits (53), Expect(2) = 9e-19
Identities = 10/25 (40%), Positives = 16/25 (64%)
Frame = -1
Query: 274 PVMLSLLPADGPTGCYFDCTEIAEF 298
PV L+LLP P+G ++ E++ F
Sbjct: 105 PVKLALLPNGSPSGLFYFRGEVSSF 31
>TC19154 similar to UP|AAS38575 (AAS38575) Short-chain dehydrogenase Tic32,
partial (44%)
Length = 508
Score = 50.4 bits (119), Expect = 4e-07
Identities = 34/86 (39%), Positives = 47/86 (54%), Gaps = 1/86 (1%)
Frame = +2
Query: 5 AVVTGANKGIGLEIVKQLAFLGVTVVLTARNDTRGRDA-ITKLHQTGLSNVMFHQLDVLD 63
AVVTGA+ GIG E + LA GV V++ RN G+D T L + + V +LD+
Sbjct: 173 AVVTGASSGIGTETTRVLAKRGVHVIMGVRNTAAGKDVKETILKENPSAKVDAMELDLSS 352
Query: 64 ALSIESLAKFIQHKFGRLDILINNAG 89
S++ A + L+ILINNAG
Sbjct: 353 MESVKKFASEYKSSGLPLNILINNAG 430
>TC17111 weakly similar to GB|AAL99238.1|22651517|AY082345 short-chain
dehydrogenase/reductase {Arabidopsis thaliana;}, partial
(53%)
Length = 561
Score = 46.6 bits (109), Expect = 5e-06
Identities = 44/169 (26%), Positives = 69/169 (40%), Gaps = 3/169 (1%)
Frame = +2
Query: 3 RYAVVTGANKGIGLEIVKQLAFLGVTVVLTARNDTRGRDAITKLHQTGLSNVMFHQLDVL 62
+ A+VTG GIG I + G + + D G L +NV F DV
Sbjct: 95 KVALVTGGASGIGESIARLFHTNGAKICIADMQDNLGNQVCESLGDE--ANVCFLHCDVT 268
Query: 63 DALSIESLAKFIQHKFGRLDILINNA---GASCVEVDKEGLKALNVDPATWLAGKVSNTL 119
+ + + KFG LDI++NNA GA C ++ NVD + + KV N
Sbjct: 269 EENDVRNAVDMTVAKFGTLDIIVNNAGISGAPCPDI-------RNVDLSDF--DKVFNVN 421
Query: 120 LQGVLTQTYKKAEECLNTNYYGVKRVTMALLPLLQLSPAKARIVNLSSL 168
++GV ++G+K ++P K I++L S+
Sbjct: 422 VKGV---------------FHGMKHAARIMIP-----KKKGSIISLCSV 508
>BP047669
Length = 544
Score = 46.2 bits (108), Expect = 7e-06
Identities = 24/87 (27%), Positives = 43/87 (48%)
Frame = +1
Query: 3 RYAVVTGANKGIGLEIVKQLAFLGVTVVLTARNDTRGRDAITKLHQTGLSNVMFHQLDVL 62
+ A++TG GIG K G V++ D G ++ K + +N+++ DV
Sbjct: 46 KVAIITGGASGIGAATAKLFVQHGAKVIIADIQDDLGM-SLCKTLEPNFNNIIYAHCDVT 222
Query: 63 DALSIESLAKFIQHKFGRLDILINNAG 89
+ +++ K+G+LDI+ NNAG
Sbjct: 223 NDSDVKNAVDMAVSKYGKLDIMYNNAG 303
>TC15724 similar to UP|Q9LHT0 (Q9LHT0) Short chain alcohol
dehydrogenase-like, partial (81%)
Length = 1014
Score = 45.4 bits (106), Expect = 1e-05
Identities = 29/88 (32%), Positives = 45/88 (50%), Gaps = 1/88 (1%)
Frame = +2
Query: 5 AVVTGANKGIGLEIVKQLAFLGVTVVLTARNDTRGRDAITKLHQTGLSNVMFHQLDVLDA 64
A+VTG +GIG +V++LA G TV +RN+ + + + G S V D
Sbjct: 128 ALVTGGTRGIGHAVVEELAEFGATVYTCSRNEVELNACLKEWQEKGFS-VSGSVCDASSP 304
Query: 65 LSIESLAKFIQHKF-GRLDILINNAGAS 91
E L + + F G+L+IL+NN G +
Sbjct: 305 PQREKLFELVASAFNGKLNILVNNVGTN 388
>TC17651 weakly similar to UP|Q8KBL5 (Q8KBL5) Oxidoreductase, short-chain
dehydrogenase/reductase family, partial (12%)
Length = 599
Score = 45.1 bits (105), Expect = 1e-05
Identities = 25/87 (28%), Positives = 45/87 (50%), Gaps = 1/87 (1%)
Frame = +1
Query: 6 VVTGANKGIGLEIVKQLAFLGVTVVLTARNDTRGRDAITKLH-QTGLSNVMFHQLDVLDA 64
++TG+ KGIG + K+ G V++ +R+ R + + G +V + DV
Sbjct: 247 LITGSTKGIGYALAKEFLKAGDNVLICSRSYERVETTVQNFRAEFGEQHVWGTKCDVRSG 426
Query: 65 LSIESLAKFIQHKFGRLDILINNAGAS 91
+++L F Q K +D+ INNAG++
Sbjct: 427 EDVKNLVSFAQEKLKYIDLWINNAGSN 507
>TC16362 similar to UP|HCD2_HUMAN (Q99714) 3-hydroxyacyl-CoA dehydrogenase
type II (Type II HADH) (Endoplasmic
reticulum-associated amyloid beta-peptide binding
protein) (Short-chain type dehydrogenase/reductase
XH98G2) , partial (7%)
Length = 613
Score = 44.7 bits (104), Expect = 2e-05
Identities = 31/90 (34%), Positives = 50/90 (55%), Gaps = 5/90 (5%)
Frame = +2
Query: 5 AVVTGANKGIGLEIVKQLAFLGVTVVLTARNDTRGRDAITKLHQTGLSNVMFHQLDV--L 62
A++TG GIGLE + L+ V V++ ARN ++A + Q S ++D+ L
Sbjct: 191 AIITGGASGIGLETARVLSLRKVHVIIAARNMNSAKEAKQLILQDNES----ARVDIMKL 358
Query: 63 DALSIESLAKFIQHKFG---RLDILINNAG 89
D S++S+ F+++ L+ILINNAG
Sbjct: 359 DLCSLKSVRSFVENFIALGLPLNILINNAG 448
>TC10829 similar to UP|Q93ZA0 (Q93ZA0) AT4g13250/F17N18_140, partial (45%)
Length = 732
Score = 33.9 bits (76), Expect(2) = 7e-05
Identities = 15/35 (42%), Positives = 22/35 (62%)
Frame = +1
Query: 57 HQLDVLDALSIESLAKFIQHKFGRLDILINNAGAS 91
H DV + ++ LA F ++ G +DI INNAGA+
Sbjct: 265 HACDVCEPRDVQRLANFAVNELGYIDIWINNAGAN 369
Score = 28.1 bits (61), Expect(2) = 7e-05
Identities = 11/46 (23%), Positives = 25/46 (53%)
Frame = +2
Query: 3 RYAVVTGANKGIGLEIVKQLAFLGVTVVLTARNDTRGRDAITKLHQ 48
R V+TG+ +G+G + ++ G V++T+R+ + +L +
Sbjct: 56 RNVVITGSTRGLGKALAREFLLSGDRVIVTSRSPESVEATVKELEE 193
>TC9483 similar to UP|Q9FK50 (Q9FK50) Brn1-like protein, partial (51%)
Length = 549
Score = 40.8 bits (94), Expect = 3e-04
Identities = 30/90 (33%), Positives = 47/90 (51%), Gaps = 3/90 (3%)
Frame = +2
Query: 3 RYAVVTGANKGIGLEIVKQLAFLGVTVVL--TARNDTRGRDAITKLHQTGLSNVMFHQLD 60
R A+VTG+++GIG EI LA LG +V+ T+ +D A + S + + D
Sbjct: 116 RVAIVTGSSRGIGREIAIHLASLGARLVINYTSNSDQANSVAAQINAGSATSRAITVKAD 295
Query: 61 VLDALSIESLAKFIQHKF-GRLDILINNAG 89
V D ++SL + F + IL+N+AG
Sbjct: 296 VSDPDQVKSLFDSAEQAFDSPVHILVNSAG 385
>TC9023 weakly similar to UP|Q9FXD7 (Q9FXD7) F12A21.14, partial (13%)
Length = 1282
Score = 39.7 bits (91), Expect = 6e-04
Identities = 37/132 (28%), Positives = 63/132 (47%), Gaps = 11/132 (8%)
Frame = +2
Query: 4 YAVVTGANKGIGLEIVKQLAFLGVTVVLTARNDTRGRDAITKLHQTGLSNVMFHQLDVLD 63
+AV+TG+ GIG + +LA G+ ++L RN T+ ++ V LD +
Sbjct: 170 WAVITGSTDGIGKAMSFELASQGLNLLLIGRNPTKLEATSKEIRDKYHVEVKILVLD-MQ 346
Query: 64 ALSIESLAKFIQHKFGRLDI--LINNAGAS------CVEVDKEGLKA---LNVDPATWLA 112
+ E + K I LD+ ++N+AG + EVD E + A +NV+ TW
Sbjct: 347 YVKGEEMGKLIVEAIDGLDVGLVVNSAGLAYPYARFLHEVDAELMDAVVKVNVEGLTW-- 520
Query: 113 GKVSNTLLQGVL 124
V+ +L G++
Sbjct: 521 --VTKAVLPGMI 550
>AV428388
Length = 369
Score = 39.3 bits (90), Expect = 8e-04
Identities = 28/84 (33%), Positives = 40/84 (47%), Gaps = 1/84 (1%)
Frame = +2
Query: 5 AVVTGANKGIGLEIVKQLAFLGVTVVLTARNDTRGRDAITKLHQTGLS-NVMFHQLDVLD 63
A++TG GIGLE + LA V++ RN + A ++ Q S V LD+
Sbjct: 116 AIITGGASGIGLETARILALRKAHVIIAVRNMVSAKKAKEQILQENESARVDIMNLDLCS 295
Query: 64 ALSIESLAKFIQHKFGRLDILINN 87
S+ S A+ L+ILINN
Sbjct: 296 VKSVRSFAENFNALDLPLNILINN 367
>BP058211
Length = 562
Score = 38.5 bits (88), Expect = 0.001
Identities = 24/88 (27%), Positives = 39/88 (44%)
Frame = +3
Query: 5 AVVTGANKGIGLEIVKQLAFLGVTVVLTARNDTRGRDAITKLHQTGLSNVMFHQLDVLDA 64
+++TGA GIG + G V++ G++ +L N F DV
Sbjct: 54 SMITGAASGIGKAAATKFINNGAKVIIADIQQKLGQETAKELGP----NATFITCDVTKE 221
Query: 65 LSIESLAKFIQHKFGRLDILINNAGASC 92
I + +F ++ +LDI+ NNAG C
Sbjct: 222 SDISNAVEFAISEYKQLDIMYNNAGVPC 305
>TC17063 weakly similar to UP|AAS50608 (AAS50608) ABL163Wp, partial (19%)
Length = 616
Score = 38.1 bits (87), Expect = 0.002
Identities = 27/92 (29%), Positives = 42/92 (45%), Gaps = 3/92 (3%)
Frame = +3
Query: 3 RYAVVTGANKGIGLEIVKQLAFLGVTVVLTARNDTRGRDAITKLHQTGLSNVMFHQLDVL 62
+ A++TG GIG EI Q G +V L R A++ L + + F V
Sbjct: 234 KVALITGGASGIGFEISTQFGKHGASVALMGRRKQVLDSAVSVLQSLSIPAIGF----VG 401
Query: 63 DALSIESLAKFIQ---HKFGRLDILINNAGAS 91
D E A+ ++ FG++DIL+N A +
Sbjct: 402 DVRKQEDAARVVESTFKHFGKIDILVNAAAGN 497
>TC16042 weakly similar to UP|Q8H0D9 (Q8H0D9) Alcohol dehydroge, partial
(94%)
Length = 965
Score = 37.4 bits (85), Expect = 0.003
Identities = 25/89 (28%), Positives = 36/89 (40%)
Frame = +2
Query: 3 RYAVVTGANKGIGLEIVKQLAFLGVTVVLTARNDTRGRDAITKLHQTGLSNVMFHQLDVL 62
+ A++TG GIG + + G VV+ D G L F +V
Sbjct: 107 KVALITGGASGIGEATARLFSKHGAQVVIADIQDDLGHSVCKDLESAS-----FVHCNVT 271
Query: 63 DALSIESLAKFIQHKFGRLDILINNAGAS 91
+E+ K G+LDI+ NNAG S
Sbjct: 272 KEDEVETAVNMAVSKHGKLDIMFNNAGIS 358
>BI419732
Length = 534
Score = 36.6 bits (83), Expect = 0.005
Identities = 30/91 (32%), Positives = 39/91 (41%), Gaps = 4/91 (4%)
Frame = +2
Query: 5 AVVTGANKGIGLEIVKQLAFLGVTVVLTARNDTRGRDAITKLHQTGLSNVMFHQLDVLDA 64
A+VTG +GIG IV LA G V +R ++ +TK Q S V D
Sbjct: 89 ALVTGGTRGIGHAIVNDLAAFGAAVHTCSRTESE----LTKCLQEWQSQGFLATGSVCDV 256
Query: 65 LSIESLAKFIQHKF----GRLDILINNAGAS 91
S K +Q G+L I INN G +
Sbjct: 257 SSRPQREKLMQEVASTFNGKLKIFINNVGTN 349
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.320 0.136 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,604,672
Number of Sequences: 28460
Number of extensions: 55974
Number of successful extensions: 277
Number of sequences better than 10.0: 57
Number of HSP's better than 10.0 without gapping: 264
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 265
length of query: 298
length of database: 4,897,600
effective HSP length: 90
effective length of query: 208
effective length of database: 2,336,200
effective search space: 485929600
effective search space used: 485929600
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 55 (25.8 bits)
Medicago: description of AC146743.8


