

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146743.14 + phase: 0
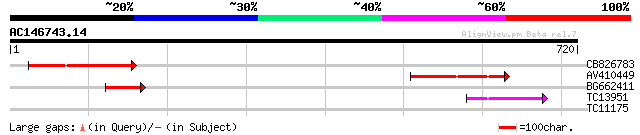
(720 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CB826783 199 2e-51
AV410449 144 4e-35
BG662411 82 2e-16
TC13951 weakly similar to UP|Q7XJQ9 (Q7XJQ9) At2g38010 protein, ... 80 9e-16
TC11175 weakly similar to GB|AAL06796.1|15809736|AY054135 AT3g60... 29 3.0
>CB826783
Length = 596
Score = 199 bits (505), Expect = 2e-51
Identities = 96/137 (70%), Positives = 118/137 (86%)
Frame = +3
Query: 25 EYLIGVGSYDMTGPAADVNMMGYANIEQNTAGIHFRLRARTFIVAENLQGPRFVFVNLDA 84
+YL+G+GSYD+TGPAADVNMMGYAN+ Q T+GIHFRL AR FIVAE +G R VFVNLDA
Sbjct: 189 DYLVGLGSYDITGPAADVNMMGYANMGQTTSGIHFRLLARAFIVAEK-KGNRVVFVNLDA 365
Query: 85 GMASQLLTIKLLERLKSRFGNLYTEENVAISGIHTHAGPGGYLQYVVYSVTSLGFVTQSF 144
MA +++TIK++ERLK+R+G+LYTE+NVAISG HTHAGPGGYLQ +VY +TS GFV QSF
Sbjct: 366 CMAPEIVTIKVIERLKARYGDLYTEKNVAISGSHTHAGPGGYLQNIVYILTSFGFVRQSF 545
Query: 145 DAIANAVEQSIIQAHNN 161
D I + +E+SI+QAH N
Sbjct: 546 DIIVDGIEKSIVQAHEN 596
>AV410449
Length = 373
Score = 144 bits (364), Expect = 4e-35
Identities = 75/125 (60%), Positives = 86/125 (68%)
Frame = +1
Query: 510 HVVIAGLTNTYSQYIATFEEYHQQRYEAASTLYGPHTLSAYIQEFNKLAQAMAKGDKIYG 569
HVV+AGLTNTYSQY+ T+EEY QRYE ASTLYGPHTLSAYIQEF KLA A+ G +
Sbjct: 7 HVVLAGLTNTYSQYVTTYEEYEVQRYEGASTLYGPHTLSAYIQEFKKLANALISGQPV-E 183
Query: 570 NGTSPPDLLSVQKSFLLDPFGDTTPDGIKLGDIKEDIAFPGSGYFTKGDKPSATFWSANP 629
G PPDLL+ Q SFL D TP G++ GD D+ P + F KGD S TFWSA P
Sbjct: 184 PGPQPPDLLNKQISFLTPVVMDGTPIGVQFGDCSSDV--PKNATFKKGDMVSVTFWSACP 357
Query: 630 RYDLL 634
R DL+
Sbjct: 358 RNDLM 372
>BG662411
Length = 503
Score = 82.4 bits (202), Expect = 2e-16
Identities = 37/51 (72%), Positives = 43/51 (83%)
Frame = +2
Query: 122 GPGGYLQYVVYSVTSLGFVTQSFDAIANAVEQSIIQAHNNLKPGSIFINTG 172
GPGG+LQYV Y VTSLGFV QSFD I + +E+SIIQAH NL+PGSIF+N G
Sbjct: 14 GPGGFLQYVTYLVTSLGFVRQSFDVIVDGIEKSIIQAHENLRPGSIFVNKG 166
>TC13951 weakly similar to UP|Q7XJQ9 (Q7XJQ9) At2g38010 protein, partial
(14%)
Length = 560
Score = 80.5 bits (197), Expect = 9e-16
Identities = 48/105 (45%), Positives = 59/105 (55%), Gaps = 2/105 (1%)
Frame = +1
Query: 581 QKSFLLDPFGDTTPDGIKLGDIKEDIAFPGSGYFTKGDKPSATFWSANPRYDLLTEGTYA 640
Q S L D TP G GD+ D+ P + F GD +A+FWSA PR DL+TEGT+A
Sbjct: 1 QISLLPPVLVDGTPFGXSFGDVCSDV--PQNSSFKTGDTVTASFWSACPRNDLMTEGTFA 174
Query: 641 VVERLQ-GERWISVQDDDDLSLFFRWKVDN-TSFHGFAAIEWEIP 683
+VE LQ + WI DDDD L F+W + S A IEW IP
Sbjct: 175 LVEFLQEKDTWIPAYDDDDFCLRFKWSRPHILSTTSKATIEWRIP 309
>TC11175 weakly similar to GB|AAL06796.1|15809736|AY054135
AT3g60690/T4C21_100 {Arabidopsis thaliana;}, partial
(42%)
Length = 692
Score = 28.9 bits (63), Expect = 3.0
Identities = 12/34 (35%), Positives = 18/34 (52%)
Frame = -3
Query: 382 NKKVVKTCPAALGPGFAAGTTDGPGVFGFQQGDP 415
NKK+ CP +G++ PG+ F+ GDP
Sbjct: 405 NKKITSACPRQS*TSINSGSSCNPGLDPFKLGDP 304
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.317 0.136 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,531,876
Number of Sequences: 28460
Number of extensions: 151021
Number of successful extensions: 705
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 697
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 701
length of query: 720
length of database: 4,897,600
effective HSP length: 97
effective length of query: 623
effective length of database: 2,136,980
effective search space: 1331338540
effective search space used: 1331338540
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 59 (27.3 bits)
Medicago: description of AC146743.14


