

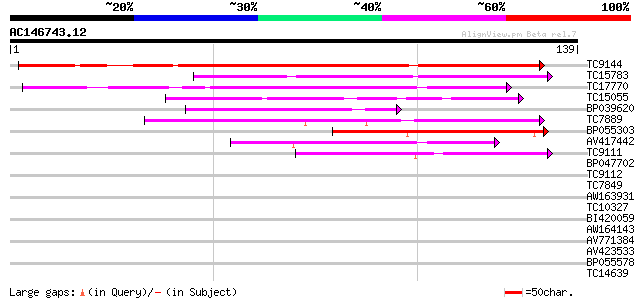
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146743.12 + phase: 0
(139 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC9144 155 3e-39
TC15783 similar to UP|Q9LJ86 (Q9LJ86) Gb|AAD48513.1 (AT3g22600/F... 79 2e-16
TC17770 weakly similar to UP|Q8VYI9 (Q8VYI9) AT5g64080/MHJ24_6, ... 67 1e-12
TC15055 weakly similar to UP|O24101 (O24101) MtN5 protein precur... 44 1e-05
BP039620 41 9e-05
TC7889 similar to UP|Q40336 (Q40336) Proline-rich cell wall prot... 41 9e-05
BP055303 41 9e-05
AV417442 40 1e-04
TC9111 39 4e-04
BP047702 37 0.001
TC9112 weakly similar to UP|Q9SI70 (Q9SI70) F23N19.16, partial (... 37 0.002
TC7849 similar to GB|AAN18126.1|23308313|BT000557 At2g10940/F15K... 36 0.003
AW163931 33 0.014
TC10327 weakly similar to UP|NLT1_PRUDU (Q43017) Nonspecific lip... 32 0.040
BI420059 31 0.089
AW164143 30 0.20
AV771384 27 0.99
AV423533 27 1.3
BP055578 27 1.7
TC14639 weakly similar to GB|AAM91068.1|22136564|AY129482 AT5g11... 26 2.2
>TC9144
Length = 835
Score = 155 bits (391), Expect = 3e-39
Identities = 84/129 (65%), Positives = 95/129 (73%)
Frame = +1
Query: 3 TTTLVVAIVSLIFASLSFHGDGVSAAALAQSPAPETAVLAPSPADDGCLMALTNMSDCLT 62
TTT VVA +L+ A L+ HG AQS APE+A APSPA C ALTN+SDCLT
Sbjct: 88 TTTRVVAFAALVLA-LACHGMS------AQSLAPESAS-APSPAGVDCFTALTNVSDCLT 243
Query: 63 FVEDGSKLTKPDKGCCPELAGLIDGNPICLCKLLGSNTADSFGIKINVNKALKLPTICGV 122
FVE GS LTKPDKGCCPE AGLI+ NPICLC+LLG D GIKIN+NKA+KLP++CGV
Sbjct: 244 FVEAGSNLTKPDKGCCPEFAGLIESNPICLCQLLGK--PDFVGIKINLNKAIKLPSVCGV 417
Query: 123 TTPPVSACS 131
TPPVS CS
Sbjct: 418 DTPPVSTCS 444
>TC15783 similar to UP|Q9LJ86 (Q9LJ86) Gb|AAD48513.1 (AT3g22600/F16J14_17),
partial (48%)
Length = 578
Score = 79.3 bits (194), Expect = 2e-16
Identities = 39/88 (44%), Positives = 50/88 (56%)
Frame = -2
Query: 46 ADDGCLMALTNMSDCLTFVEDGSKLTKPDKGCCPELAGLIDGNPICLCKLLGSNTADSFG 105
A GC ALT++S CL ++ S + P CC +L+ ++ +P CLC LL SFG
Sbjct: 373 AQSGCTSALTSLSPCLNYITGSS--SSPSPSCCSQLSSVVQSSPQCLCSLLNGG-GSSFG 203
Query: 106 IKINVNKALKLPTICGVTTPPVSACSGK 133
I +N AL LP C V TPPVS C GK
Sbjct: 202 ITMNQTLALSLPGPCKVQTPPVSQCKGK 119
>TC17770 weakly similar to UP|Q8VYI9 (Q8VYI9) AT5g64080/MHJ24_6, partial
(42%)
Length = 554
Score = 66.6 bits (161), Expect = 1e-12
Identities = 41/120 (34%), Positives = 65/120 (54%)
Frame = +3
Query: 4 TTLVVAIVSLIFASLSFHGDGVSAAALAQSPAPETAVLAPSPADDGCLMALTNMSDCLTF 63
++L+ + VSLI ++ + A A S + A P+P+ D C + M+DCL+F
Sbjct: 165 SSLMASKVSLILCVVA-----ICAVDFAHSASSNNA---PAPSAD-CSNLILKMTDCLSF 317
Query: 64 VEDGSKLTKPDKGCCPELAGLIDGNPICLCKLLGSNTADSFGIKINVNKALKLPTICGVT 123
V + S +TKP CC L ++ P CLC+ S++ FG+ +N KAL LP+ C V+
Sbjct: 318 VTNDSTVTKPQGQCCSGLKSVLKTAPSCLCEAFKSSS--QFGVVLNATKALALPSACKVS 491
>TC15055 weakly similar to UP|O24101 (O24101) MtN5 protein precursor,
partial (52%)
Length = 664
Score = 43.5 bits (101), Expect = 1e-05
Identities = 31/88 (35%), Positives = 42/88 (47%)
Frame = +3
Query: 39 AVLAPSPADDGCLMALTNMSDCLTFVEDGSKLTKPDKGCCPELAGLIDGNPICLCKLLGS 98
A+L + A C + + + C G PDK CC + N CLCK
Sbjct: 93 ALLGGAQAVALCNIDTSQLKSCRA-AATGEHPPPPDKKCCDVVR---QANLPCLCKY--K 254
Query: 99 NTADSFGIKINVNKALKLPTICGVTTPP 126
+ SFGI N +ALKLP+ CG++TPP
Sbjct: 255 SALPSFGI--NPTQALKLPSECGLSTPP 332
>BP039620
Length = 503
Score = 40.8 bits (94), Expect = 9e-05
Identities = 21/53 (39%), Positives = 27/53 (50%)
Frame = +3
Query: 44 SPADDGCLMALTNMSDCLTFVEDGSKLTKPDKGCCPELAGLIDGNPICLCKLL 96
S A + C LTN++ C FV G+ TKP CC L+ + N CLC L
Sbjct: 117 SEAQNSCTNQLTNLNVCAPFVVPGASNTKPSADCCGALSAV---NHECLCSTL 266
>TC7889 similar to UP|Q40336 (Q40336) Proline-rich cell wall protein,
partial (66%)
Length = 1158
Score = 40.8 bits (94), Expect = 9e-05
Identities = 30/102 (29%), Positives = 43/102 (41%), Gaps = 4/102 (3%)
Frame = +1
Query: 34 PAPETAVLAPSPADDGCLMALTNMSDCLTFVEDGSKLT---KPDKGCCPELAGLID-GNP 89
P P V +P PA C + + C+ + + + CCP LAGL+D
Sbjct: 487 PPPPPVVPSPPPAQPTCPIDTLKLGACVDLLGGLIHIGIGGSAKQTCCPVLAGLVDLDAA 666
Query: 90 ICLCKLLGSNTADSFGIKINVNKALKLPTICGVTTPPVSACS 131
+CLC + A I I + AL+L CG T P C+
Sbjct: 667 VCLCTTI---RAKLLNINIIIPIALQLLLDCGKTPPDGFKCA 783
>BP055303
Length = 515
Score = 40.8 bits (94), Expect = 9e-05
Identities = 19/55 (34%), Positives = 35/55 (63%), Gaps = 2/55 (3%)
Frame = -1
Query: 80 ELAGLIDGNPICLCKLL-GSNTADSFGIKINVNKALKLPTICGVTTPPV-SACSG 132
+L+ ++ P CLC+++ G ++ + + IN +AL LP+ C V TPP+ + C+G
Sbjct: 512 QLSFVVKSQPQCLCEVVNGGASSIAASLNINQTQALTLPSACNVQTPPITTTCTG 348
>AV417442
Length = 391
Score = 40.4 bits (93), Expect = 1e-04
Identities = 22/67 (32%), Positives = 32/67 (46%), Gaps = 1/67 (1%)
Frame = +3
Query: 55 TNMSDCLTFVEDGS-KLTKPDKGCCPELAGLIDGNPICLCKLLGSNTADSFGIKINVNKA 113
T+ + C++F+ + S T P CC + L G CLC ++ N F I IN A
Sbjct: 195 TSFTPCMSFLTNSSGNGTSPTAECCDSIKSLTSGGRDCLCLVVTGNV--PFSIPINRTLA 368
Query: 114 LKLPTIC 120
+ LP C
Sbjct: 369 ISLPRAC 389
>TC9111
Length = 534
Score = 38.5 bits (88), Expect = 4e-04
Identities = 24/64 (37%), Positives = 32/64 (49%), Gaps = 1/64 (1%)
Frame = +2
Query: 71 TKPDKGCCPELAGLIDGNPICLCKLLGS-NTADSFGIKINVNKALKLPTICGVTTPPVSA 129
T P CC + ++ CLC L S SF KI+ ++AL+L CGVT+ S
Sbjct: 197 TNPPASCCDAIKKTVETQLTCLCNLFYSPGLLQSF--KISTDQALQLSRRCGVTSDLSSC 370
Query: 130 CSGK 133
SGK
Sbjct: 371 KSGK 382
>BP047702
Length = 391
Score = 37.4 bits (85), Expect = 0.001
Identities = 22/74 (29%), Positives = 32/74 (42%)
Frame = +3
Query: 57 MSDCLTFVEDGSKLTKPDKGCCPELAGLIDGNPICLCKLLGSNTADSFGIKINVNKALKL 116
+S C FV+ P CC L + CLCK + +I++ KA+ +
Sbjct: 84 VSQCQAFVKKEGPQILPSDACCETLKA---ADIPCLCKYI----TPVIQSQISIEKAIYV 242
Query: 117 PTICGVTTPPVSAC 130
CG T PPV+ C
Sbjct: 243 VKTCGGTVPPVTKC 284
>TC9112 weakly similar to UP|Q9SI70 (Q9SI70) F23N19.16, partial (53%)
Length = 807
Score = 36.6 bits (83), Expect = 0.002
Identities = 23/63 (36%), Positives = 31/63 (48%), Gaps = 1/63 (1%)
Frame = +1
Query: 71 TKPDKGCCPELAGLIDGNPICLCKLLGS-NTADSFGIKINVNKALKLPTICGVTTPPVSA 129
T P CC + ++ CLC L S SF KI+ ++AL+L CGVT+ S
Sbjct: 217 TNPPASCCDAIKKTVETQLTCLCNLFYSPGLLQSF--KISTDQALQLSRRCGVTSDLSSC 390
Query: 130 CSG 132
SG
Sbjct: 391 KSG 399
>TC7849 similar to GB|AAN18126.1|23308313|BT000557 At2g10940/F15K19.1
{Arabidopsis thaliana;}, partial (55%)
Length = 1157
Score = 35.8 bits (81), Expect = 0.003
Identities = 22/56 (39%), Positives = 27/56 (47%), Gaps = 1/56 (1%)
Frame = +2
Query: 77 CCPELAGLIDGNPI-CLCKLLGSNTADSFGIKINVNKALKLPTICGVTTPPVSACS 131
CCP L GL++ CLC L + + I V AL+L CG T PP CS
Sbjct: 605 CCPVLQGLVEVEAAACLCTTLKLKLLN---LNIYVPLALQLLVACGKTPPPGYTCS 763
>AW163931
Length = 392
Score = 33.5 bits (75), Expect = 0.014
Identities = 18/53 (33%), Positives = 26/53 (48%)
Frame = +2
Query: 75 KGCCPELAGLIDGNPICLCKLLGSNTADSFGIKINVNKALKLPTICGVTTPPV 127
+ CC ++ NP CLC +L SNTA GI + A+ +P C P+
Sbjct: 206 QSCCAQVKKFXQ-NPGCLCAVLXSNTAKMSGIDPXI--AITIPKRCSFANRPI 355
>TC10327 weakly similar to UP|NLT1_PRUDU (Q43017) Nonspecific lipid-transfer
protein 1 precursor (LTP 1), partial (52%)
Length = 683
Score = 32.0 bits (71), Expect = 0.040
Identities = 26/94 (27%), Positives = 40/94 (41%), Gaps = 12/94 (12%)
Frame = +1
Query: 50 CLMALTNMSDCLTFVEDGSKLTKPDKGCCPELAGLI-------DGNPICLC-KLLGSNTA 101
C + ++ C++++ +G + CC + L D +C C K SN+
Sbjct: 166 CGQVVNSLYPCVSYLMNGGNMVPGQ--CCNGIRNLYGMATNTPDRRAVCTCIKQAVSNSG 339
Query: 102 DSFGIKINVNKALKLPTICGVTTP----PVSACS 131
SF N+N A LP CGV P P + CS
Sbjct: 340 FSF-TSYNLNLAAGLPKKCGVNIPYQISPNTDCS 438
>BI420059
Length = 461
Score = 30.8 bits (68), Expect = 0.089
Identities = 21/61 (34%), Positives = 29/61 (47%), Gaps = 2/61 (3%)
Frame = +3
Query: 73 PDKGCCPELAGLID-GNPICLCKLLGSNTADSFGIKINVNKALK-LPTICGVTTPPVSAC 130
P K CC L GL+D +CLC L +N GI +N+ +L L +C P C
Sbjct: 33 PVKPCCTLLNGLVDLEAAVCLCTALKANI---LGINLNLPISLSLLLNVCSKQVPRDFQC 203
Query: 131 S 131
+
Sbjct: 204 A 206
>AW164143
Length = 412
Score = 29.6 bits (65), Expect = 0.20
Identities = 14/22 (63%), Positives = 18/22 (81%)
Frame = +2
Query: 25 VSAAALAQSPAPETAVLAPSPA 46
VSAAA A +PAP +A +AP+PA
Sbjct: 68 VSAAAPAPAPAPVSASIAPAPA 133
>AV771384
Length = 359
Score = 27.3 bits (59), Expect = 0.99
Identities = 11/18 (61%), Positives = 13/18 (72%), Gaps = 1/18 (5%)
Frame = -3
Query: 77 CCPELAGLID-GNPICLC 93
CCP LAGL+D +CLC
Sbjct: 336 CCPVLAGLVDLDAAVCLC 283
>AV423533
Length = 472
Score = 26.9 bits (58), Expect = 1.3
Identities = 19/64 (29%), Positives = 27/64 (41%), Gaps = 4/64 (6%)
Frame = -2
Query: 15 FASLSFHGDGVSAAALAQSPAPETAVLAPSPADDGCLMA----LTNMSDCLTFVEDGSKL 70
F SL+FH + + P V P PA D ++A N +D ++ GS L
Sbjct: 324 FPSLAFHRAAGGYGGIKIASRPLIPVPFPPPAGDFTILAGDWYKRNHTDLRAILDSGSDL 145
Query: 71 TKPD 74
PD
Sbjct: 144 PFPD 133
>BP055578
Length = 506
Score = 26.6 bits (57), Expect = 1.7
Identities = 18/71 (25%), Positives = 27/71 (37%), Gaps = 3/71 (4%)
Frame = +1
Query: 49 GCLMALTN-MSDCLTFVED--GSKLTKPDKGCCPELAGLIDGNPICLCKLLGSNTADSFG 105
GC L + S CL + ++ CC + N +C C+LL A G
Sbjct: 97 GCADELMSPFSPCLPYFSSPPNNRTETASGACCHAFSSATTVNSLCFCQLL--RDASILG 270
Query: 106 IKINVNKALKL 116
+N + L L
Sbjct: 271 FPLNTTRLLSL 303
>TC14639 weakly similar to GB|AAM91068.1|22136564|AY129482
AT5g11740/T22P22_130 {Arabidopsis thaliana;}, partial
(61%)
Length = 507
Score = 26.2 bits (56), Expect = 2.2
Identities = 17/52 (32%), Positives = 31/52 (58%)
Frame = +1
Query: 2 ATTTLVVAIVSLIFASLSFHGDGVSAAALAQSPAPETAVLAPSPADDGCLMA 53
+T +++A +S++ A +S V+AAA A +P+P + A SP+ L+A
Sbjct: 70 STRVMLLATMSVLAAVIS-----VAAAAEAPAPSPASPASAVSPSFAAGLLA 210
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.320 0.135 0.406
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,700,076
Number of Sequences: 28460
Number of extensions: 39339
Number of successful extensions: 235
Number of sequences better than 10.0: 49
Number of HSP's better than 10.0 without gapping: 229
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 229
length of query: 139
length of database: 4,897,600
effective HSP length: 81
effective length of query: 58
effective length of database: 2,592,340
effective search space: 150355720
effective search space used: 150355720
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 50 (23.9 bits)
Medicago: description of AC146743.12


