

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
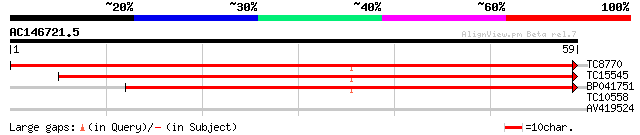
Query= AC146721.5 - phase: 0 /pseudo
(59 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC8770 homologue to UP|O22111 (O22111) 6-phosphogluconate dehydr... 73 1e-14
TC15545 homologue to GB|AAN73296.1|25141203|BT002299 At3g02360/F... 72 2e-14
BP041751 47 1e-06
TC10558 similar to UP|LBD6_ARATH (O04479) LOB domain protein 6 (... 24 5.2
AV419524 23 8.9
>TC8770 homologue to UP|O22111 (O22111) 6-phosphogluconate dehydrogenase,
decarboxylating , partial (70%)
Length = 1154
Score = 72.8 bits (177), Expect = 1e-14
Identities = 39/69 (56%), Positives = 44/69 (63%), Gaps = 10/69 (14%)
Frame = +3
Query: 1 MATSEPTNLARIGLAVIGQNLALNIAEKGFQFQF----------TIERAKQEGNLPIYGY 50
MA T + GLAV+GQNLALNIAEKGF T+ERAKQEGNLP+YGY
Sbjct: 51 MAQPNLTRIGLAGLAVMGQNLALNIAEKGFPISVYNRTTSKVDETVERAKQEGNLPVYGY 230
Query: 51 HDFKYFVQS 59
HD + FV S
Sbjct: 231 HDAEAFVHS 257
>TC15545 homologue to GB|AAN73296.1|25141203|BT002299 At3g02360/F11A12_104
{Arabidopsis thaliana;} , partial (31%)
Length = 663
Score = 72.4 bits (176), Expect = 2e-14
Identities = 37/64 (57%), Positives = 44/64 (67%), Gaps = 10/64 (15%)
Frame = +1
Query: 6 PTNLARIGLAVIGQNLALNIAEKGFQFQF----------TIERAKQEGNLPIYGYHDFKY 55
PT + GLAV+GQNLALNIA+KGF T+ERAK+EGNLP+YGYHD +
Sbjct: 208 PTRIGLAGLAVMGQNLALNIADKGFPISVYNRTTSKVDETVERAKKEGNLPVYGYHDPES 387
Query: 56 FVQS 59
FVQS
Sbjct: 388 FVQS 399
>BP041751
Length = 471
Score = 46.6 bits (109), Expect = 1e-06
Identities = 26/57 (45%), Positives = 36/57 (62%), Gaps = 10/57 (17%)
Frame = +3
Query: 13 GLAVIGQNLALNIAEKGFQFQF----------TIERAKQEGNLPIYGYHDFKYFVQS 59
GLAV+GQ LALNIA+KGF T++RA +EG+LP+ G ++ + FV S
Sbjct: 165 GLAVMGQXLALNIADKGFPISVYNRTTSKVDETVDRAHREGSLPLIGQYNPRDFVLS 335
>TC10558 similar to UP|LBD6_ARATH (O04479) LOB domain protein 6 (ASYMMETRIC
LEAVES2), partial (20%)
Length = 669
Score = 24.3 bits (51), Expect = 5.2
Identities = 11/26 (42%), Positives = 16/26 (61%)
Frame = -3
Query: 1 MATSEPTNLARIGLAVIGQNLALNIA 26
+A +E + + GLA+ GQNLA A
Sbjct: 607 LAKNELSKIQNFGLAMAGQNLAATTA 530
>AV419524
Length = 194
Score = 23.5 bits (49), Expect = 8.9
Identities = 10/19 (52%), Positives = 12/19 (62%)
Frame = +1
Query: 2 ATSEPTNLARIGLAVIGQN 20
A S P L R+GL IG+N
Sbjct: 70 AVSLPAKLCRLGLTYIGKN 126
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.321 0.138 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 729,293
Number of Sequences: 28460
Number of extensions: 5348
Number of successful extensions: 18
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 16
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 16
length of query: 59
length of database: 4,897,600
effective HSP length: 35
effective length of query: 24
effective length of database: 3,901,500
effective search space: 93636000
effective search space used: 93636000
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 49 (23.5 bits)
Medicago: description of AC146721.5


