

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146709.2 - phase: 0
(751 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
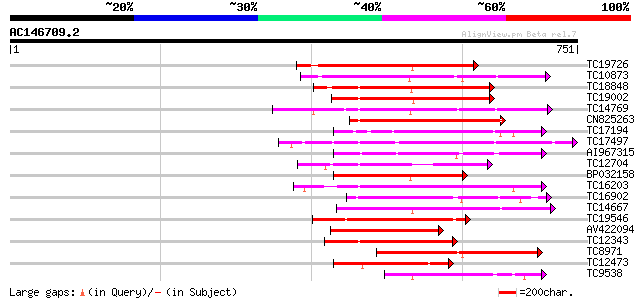
Score E
Sequences producing significant alignments: (bits) Value
TC19726 similar to UP|Q9XIS3 (Q9XIS3) Lectin-like protein kinase... 222 1e-58
TC10873 similar to UP|Q9LDZ5 (Q9LDZ5) F2D10.13 (F5M15.3), partia... 213 1e-55
TC18848 weakly similar to UP|Q9SSQ6 (Q9SSQ6) F6D8.24 protein, pa... 207 5e-54
TC19002 weakly similar to UP|Q9XJ10 (Q9XJ10) ESTs C22458(C62866)... 201 5e-52
TC14769 UP|Q8LKX1 (Q8LKX1) Receptor-like kinase SYMRK, complete 200 6e-52
CN825263 190 8e-49
TC17194 UP|CAE02590 (CAE02590) Nod-factor receptor 1b, complete 189 2e-48
TC17497 UP|CAE45595 (CAE45595) S-receptor kinase-like protein 2,... 183 8e-47
AI967315 177 5e-45
TC12704 UP|CAE45596 (CAE45596) S-receptor kinase-like protein 3,... 170 7e-43
BP032158 168 3e-42
TC16203 UP|Q8GRU6 (Q8GRU6) LRR receptor-like kinase (Hypernodula... 166 1e-41
TC16902 homologue to UP|Q8VZH5 (Q8VZH5) Receptor protein kinase-... 166 1e-41
TC14667 homologue to UP|Q84P43 (Q84P43) Protein kinase Pti1, com... 162 1e-40
TC19546 similar to UP|Q40543 (Q40543) Protein-serine/threonine k... 160 7e-40
AV422094 158 3e-39
TC12343 similar to UP|CDK9_SCHPO (Q96WV9) Serine/threonine prote... 158 3e-39
TC8971 similar to GB|AAM16251.1|20334780|AY093990 At2g11520/F14P... 153 1e-37
TC12473 similar to UP|Q9M068 (Q9M068) Protein kinase-like protei... 152 2e-37
TC9538 similar to UP|RLK5_ARATH (P47735) Receptor-like protein k... 146 1e-35
>TC19726 similar to UP|Q9XIS3 (Q9XIS3) Lectin-like protein kinase, partial
(30%)
Length = 728
Score = 222 bits (566), Expect = 1e-58
Identities = 117/249 (46%), Positives = 165/249 (65%), Gaps = 8/249 (3%)
Frame = +3
Query: 380 LGAVIIGV-GALVAYVILKRKRKRSEKQKEEAMHLTSMNDDLERGAGPRRFTYKELELAT 438
LG VI+ V G L+ ++ LK+KR EE + + PR F Y EL+ AT
Sbjct: 3 LGFVIVAVIGGLLWWIRLKKKR-------EEGSNSQILGTLKSLPGTPREFNYVELKKAT 161
Query: 439 NNFSKDRKLGQGGFGAVFKGYFADLDLQVAVKKISRGSRQGKKEYVTEVKVISQLRHRNL 498
NNF + KLGQGG+G V++G L+VAVK SR + ++++E+ +I++LRH++L
Sbjct: 162 NNFDEKHKLGQGGYGVVYRGMLPKEKLEVAVKMFSRDKMKSTDDFLSELIIINRLRHKHL 341
Query: 499 VKLLGWCHDKGEFLLIYEFMPNGSLDSHLFGKR-----TPLSWGVRHKITLGLASGLLYL 553
V+L GWCH G LL+Y++MPNGSLDSH+F + TPLSW +R+KI G+AS L YL
Sbjct: 342 VRLQGWCHKNGVLLLVYDYMPNGSLDSHIFCEEGGTITTPLSWPLRYKIISGVASALNYL 521
Query: 554 HEEWERCVVHRDIKSSNVMLDSSFNVKLGDFGLAKLMDHELGPQT--TGLAGTFGYLAPE 611
H E+++ VVHRD+K+SN+MLDS FN KLGDFGLA+ +++E T G+ GT GY+APE
Sbjct: 522 HNEYDQKVVHRDLKASNIMLDSEFNAKLGDFGLARALENEKISYTELEGVHGTMGYIAPE 701
Query: 612 YVSTGRASK 620
TG+A++
Sbjct: 702 CFHTGKATR 728
>TC10873 similar to UP|Q9LDZ5 (Q9LDZ5) F2D10.13 (F5M15.3), partial (77%)
Length = 1478
Score = 213 bits (541), Expect = 1e-55
Identities = 126/338 (37%), Positives = 191/338 (56%), Gaps = 7/338 (2%)
Frame = +1
Query: 386 GVGALVAYVILKRKRKRSEKQKEEAMHLTSMNDDLERGAGPRRFTYKELELATNNFSKDR 445
GV A A +RKR+ K K ++S N F ++EL AT NF +
Sbjct: 298 GVNAAAAATDGERKRETKGKGKG----VSSSNGSSNGKTAAASFGFRELADATRNFKEAN 465
Query: 446 KLGQGGFGAVFKGYFADLDLQVAVKKISRGSRQGKKEYVTEVKVISQLRHRNLVKLLGWC 505
+G+GGFG V+KG + VAVK++S RQG +E+V EV ++S L H NLV+L+G+C
Sbjct: 466 LIGEGGFGKVYKGRLTTGEA-VAVKQLSHDGRQGFQEFVMEVLMLSLLHHTNLVRLIGYC 642
Query: 506 HDKGEFLLIYEFMPNGSLDSHLF---GKRTPLSWGVRHKITLGLASGLLYLHEEWERCVV 562
D + LL+YE+MP GSL+ HLF + PL+W R K+ +G A GL YLH + V+
Sbjct: 643 TDGDQRLLVYEYMPMGSLEDHLFELSHDKEPLNWSTRMKVAVGAARGLEYLHCTADPPVI 822
Query: 563 HRDIKSSNVMLDSSFNVKLGDFGLAKLMDHELGPQT---TGLAGTFGYLAPEYVSTGRAS 619
+RD+KS+N++LD+ FN KL DFGLAKL +G T T + GT+GY APEY +G+ +
Sbjct: 823 YRDLKSANILLDNEFNPKLSDFGLAKL--GPVGDNTHVSTRVMGTYGYCAPEYAMSGKLT 996
Query: 620 KESDVYSFGIVVLEITSGKKATEVMKEKDEEKGMIEWVWDHYGKGELLVAM-DENLRKDF 678
+SD+YSFG+V+LE+ +G++A + + + E+ ++ W ++ M D L+ F
Sbjct: 997 LKSDIYSFGVVLLELLTGRRAIDTSR-RPGEQNLVSWARPYFSDRRRFGHMVDPLLQGRF 1173
Query: 679 DEKQVECLMIVGLWCAHPDVSLRPSIRQAIQVLNFEVS 716
+ + + + C RP I + L + S
Sbjct: 1174PSRCLHQAIAITAMCLQEQPKFRPLITDIVVALEYLAS 1287
>TC18848 weakly similar to UP|Q9SSQ6 (Q9SSQ6) F6D8.24 protein, partial (64%)
Length = 969
Score = 207 bits (527), Expect = 5e-54
Identities = 111/243 (45%), Positives = 156/243 (63%), Gaps = 3/243 (1%)
Frame = +2
Query: 403 SEKQKEEAMHLTSMNDDLERGAGPRRFTYKELELATNNFSKDRKLGQGGFGAVFKGYFAD 462
SEK +E S+N+ R FTYKEL AT FS D KLG+GGFG+V+ G +D
Sbjct: 206 SEKVEEGPTSFGSVNNSW------RIFTYKELHAATGGFSDDNKLGEGGFGSVYWGRTSD 367
Query: 463 LDLQVAVKKISRGSRQGKKEYVTEVKVISQLRHRNLVKLLGWCHDKGEFLLIYEFMPNGS 522
LQ+AVKK+ + + + E+ EV+V+ ++RH+NL+ L G+C + L++Y++MPN S
Sbjct: 368 -GLQIAVKKLKAMNSKAEMEFAVEVEVLGRVRHKNLLGLRGYCVGDDQRLIVYDYMPNLS 544
Query: 523 LDSHLFGK---RTPLSWGVRHKITLGLASGLLYLHEEWERCVVHRDIKSSNVMLDSSFNV 579
L SHL G+ L+W R KI +G A G+LYLH E ++HRDIK+SNV+L+S F
Sbjct: 545 LLSHLHGQFAVEVQLNWQKRMKIAIGSAEGILYLHHEVTPHIIHRDIKASNVLLNSDFEP 724
Query: 580 KLGDFGLAKLMDHELGPQTTGLAGTFGYLAPEYVSTGRASKESDVYSFGIVVLEITSGKK 639
+ DFG AKL+ + TT + GT GYLAPEY G+ S+ DVYSFGI++LE+ +G+K
Sbjct: 725 LVADFGFAKLIPEGVSHMTTRVKGTLGYLAPEYAMWGKVSESCDVYSFGILLLELVTGRK 904
Query: 640 ATE 642
E
Sbjct: 905 PIE 913
>TC19002 weakly similar to UP|Q9XJ10 (Q9XJ10) ESTs C22458(C62866), partial
(48%)
Length = 780
Score = 201 bits (510), Expect = 5e-52
Identities = 102/219 (46%), Positives = 147/219 (66%), Gaps = 3/219 (1%)
Frame = +1
Query: 427 RRFTYKELELATNNFSKDRKLGQGGFGAVFKGYFADLDLQVAVKKISRGSRQGKKEYVTE 486
R F+ KEL ATNNF+ D KLG+GGFG+V+ G D Q+AVK++ S + E+ E
Sbjct: 22 RVFSLKELHSATNNFNYDNKLGEGGFGSVYWGQLWD-GSQIAVKRLKVWSNKADMEFAVE 198
Query: 487 VKVISQLRHRNLVKLLGWCHDKGEFLLIYEFMPNGSLDSHLFGKRTP---LSWGVRHKIT 543
V++++++RH+NL+ L G+C + E L++Y++MPN SL SHL G+ + L W R I
Sbjct: 199 VEILARVRHKNLLSLRGYCAEGQERLIVYDYMPNLSLLSHLHGQHSSECLLDWNRRMNIA 378
Query: 544 LGLASGLLYLHEEWERCVVHRDIKSSNVMLDSSFNVKLGDFGLAKLMDHELGPQTTGLAG 603
+G A G++YLH + ++HRDIK+SNV+LDS F ++ DFG AKL+ TT + G
Sbjct: 379 IGSAEGIVYLHHQATPHIIHRDIKASNVLLDSDFQARVADFGFAKLIPDGATHVTTRVKG 558
Query: 604 TFGYLAPEYVSTGRASKESDVYSFGIVVLEITSGKKATE 642
T GYLAPEY G+A++ DV+SFGI++LE+ SGKK E
Sbjct: 559 TLGYLAPEYAMLGKANECCDVFSFGILLLELASGKKPLE 675
>TC14769 UP|Q8LKX1 (Q8LKX1) Receptor-like kinase SYMRK, complete
Length = 3495
Score = 200 bits (509), Expect = 6e-52
Identities = 128/385 (33%), Positives = 207/385 (53%), Gaps = 14/385 (3%)
Frame = +3
Query: 349 NLLSWEFNSNLEKSDDSNSKDTRLVVILTVSLGAVIIGVGALVAYVILKRKR-------- 400
N+ S N++ + S+ +++VI ++ G+++I + V +V R++
Sbjct: 1866 NMNSSLINTDYGRCKGKESRFGQVIVIGAITCGSLLITLAFGVLFVCRYRQKLIPWEGFA 2045
Query: 401 -KRSEKQKEEAMHLTSMNDDLERGAGPRRFTYKELELATNNFSKDRKLGQGGFGAVFKGY 459
K+ + L S +D + + FT + +E+AT + +G+GGFG+V++G
Sbjct: 2046 GKKYPMETNIIFSLPSKDDFFIKSVSIQAFTLEYIEVATERYKT--LIGEGGFGSVYRGT 2219
Query: 460 FADLDLQVAVKKISRGSRQGKKEYVTEVKVISQLRHRNLVKLLGWCHDKGEFLLIYEFMP 519
D +VAVK S S QG +E+ E+ ++S ++H NLV LLG+C++ + +L+Y FM
Sbjct: 2220 LND-GQEVAVKVRSSTSTQGTREFDNELNLLSAIQHENLVPLLGYCNESDQQILVYPFMS 2396
Query: 520 NGSLDSHLFG---KRTPLSWGVRHKITLGLASGLLYLHEEWERCVVHRDIKSSNVMLDSS 576
NGSL L+G KR L W R I LG A GL YLH R V+HRDIKSSN++LD S
Sbjct: 2397 NGSLQDRLYGEPAKRKILDWPTRLSIALGAARGLAYLHTFPGRSVIHRDIKSSNILLDHS 2576
Query: 577 FNVKLGDFGLAKLMDHELGPQTTGL--AGTFGYLAPEYVSTGRASKESDVYSFGIVVLEI 634
K+ DFG +K E G L GT GYL PEY T + S++SDV+SFG+V+LEI
Sbjct: 2577 MCAKVADFGFSKYAPQE-GDSYVSLEVRGTAGYLDPEYYKTQQLSEKSDVFSFGVVLLEI 2753
Query: 635 TSGKKATEVMKEKDEEKGMIEWVWDHYGKGELLVAMDENLRKDFDEKQVECLMIVGLWCA 694
SG++ + K E ++EW + ++ +D ++ + + + ++ V L C
Sbjct: 2754 VSGREPLNI-KRPRTEWSLVEWATPYIRGSKVDEIVDPGIKGGYHAEAMWRVVEVALQCL 2930
Query: 695 HPDVSLRPSIRQAIQVLNFEVSMPN 719
P + RPS+ ++ L + + N
Sbjct: 2931 EPFSTYRPSMVAIVRELEDALIIEN 3005
>CN825263
Length = 663
Score = 190 bits (482), Expect = 8e-49
Identities = 101/210 (48%), Positives = 139/210 (66%), Gaps = 4/210 (1%)
Frame = +1
Query: 451 GFGAVFKGYFADLDLQVAVKKISRGSRQGKKEYVTEVKVISQLRHRNLVKLLGWCHDKGE 510
GFG V+KG D VAVK + R ++G +E++ EV+++S+L HRNLVKL+G C +K
Sbjct: 1 GFGLVYKGILND-GRDVAVKILKRDDQRGGREFLAEVEMLSRLHHRNLVKLIGICIEKQT 177
Query: 511 FLLIYEFMPNGSLDSHLFG--KRT-PLSWGVRHKITLGLASGLLYLHEEWERCVVHRDIK 567
LIYE +PNGS++SHL G K T PL W R KI LG A GL YLHE+ CV+HRD K
Sbjct: 178 RCLIYELVPNGSVESHLHGADKETGPLDWNARMKIALGAARGLAYLHEDSNPCVIHRDFK 357
Query: 568 SSNVMLDSSFNVKLGDFGLAK-LMDHELGPQTTGLAGTFGYLAPEYVSTGRASKESDVYS 626
SSN++L+ F K+ DFGLA+ +D +T + GTFGYLAPEY TG +SDVYS
Sbjct: 358 SSNILLECDFTPKVSDFGLARTALDEGNKHISTHVMGTFGYLAPEYAMTGHLLVKSDVYS 537
Query: 627 FGIVVLEITSGKKATEVMKEKDEEKGMIEW 656
+G+V+LE+ +G K ++ + +E ++ W
Sbjct: 538 YGVVLLELLTGTKPVDLSQPPGQE-NLVTW 624
>TC17194 UP|CAE02590 (CAE02590) Nod-factor receptor 1b, complete
Length = 2193
Score = 189 bits (479), Expect = 2e-48
Identities = 113/292 (38%), Positives = 169/292 (57%), Gaps = 9/292 (3%)
Frame = +1
Query: 429 FTYKELELATNNFSKDRKLGQGGFGAVFKGYFADL-DLQVAVKKISRGSRQGKKEYVTEV 487
F+Y+EL ATNNFS D K+GQGGFGAV Y+A+L + A+KK+ Q E++ E+
Sbjct: 1051 FSYQELAKATNNFSLDNKIGQGGFGAV---YYAELRGKKTAIKKMDV---QASTEFLCEL 1212
Query: 488 KVISQLRHRNLVKLLGWCHDKGEFLLIYEFMPNGSLDSHLFGK-RTPLSWGVRHKITLGL 546
KV++ + H NLV+L+G+C + G L+YE + NG+L +L G + PL W R +I L
Sbjct: 1213 KVLTHVHHLNLVRLIGYCVE-GSLFLVYEHIDNGNLGQYLHGSGKEPLPWSSRVQIALDA 1389
Query: 547 ASGLLYLHEEWERCVVHRDIKSSNVMLDSSFNVKLGDFGLAKLMDHELGPQTTGLAGTFG 606
A GL Y+HE +HRD+KS+N+++D + K+ DFGL KL++ T L GTFG
Sbjct: 1390 ARGLEYIHEHTVPVYIHRDVKSANILIDKNLRGKVADFGLTKLIEVGNSTLQTRLVGTFG 1569
Query: 607 YLAPEYVSTGRASKESDVYSFGIVVLEITSGKKATEVMKEKD---EEKGMIEWVWDHYGK 663
Y+ PEY G S + DVY+FG+V+ E+ S K A V+K + E KG++ + K
Sbjct: 1570 YMPPEYAQYGDISPKIDVYAFGVVLFELISAKNA--VLKTGELVAESKGLVALFEEALNK 1743
Query: 664 GE----LLVAMDENLRKDFDEKQVECLMIVGLWCAHPDVSLRPSIRQAIQVL 711
+ L +D L +++ V + +G C + LRPS+R + L
Sbjct: 1744 SDPCDALRKLVDPRLGENYPIDSVLKIAQLGRACTRDNPLLRPSMRSLVVAL 1899
>TC17497 UP|CAE45595 (CAE45595) S-receptor kinase-like protein 2, complete
Length = 2946
Score = 183 bits (465), Expect = 8e-47
Identities = 134/407 (32%), Positives = 212/407 (51%), Gaps = 12/407 (2%)
Frame = +1
Query: 357 SNLEKSDDSNSKDTR-----LVVILTVSLGAVIIGVGALVAYVILKRKRKRSEKQKEEAM 411
S L+++ + S +T+ LVVI+ + I+G+ ++ I ++K KR ++ + +
Sbjct: 1411 SKLDRTRNKKSINTKKLAGSLVVIIAFVIFITILGLA--ISTCIQRKKNKRGDEGEIGII 1584
Query: 412 -HLTSMNDDLERGAGPRRFTYKELELATNNFSKDRKLGQGGFGAVFKGYFADLDLQVAVK 470
H D + F + + ATN+FS KLG+GGFG V+KG A+ ++AVK
Sbjct: 1585 NHWKDKRGDEDIDLATI-FDFSTISSATNHFSLSNKLGEGGFGPVYKGLLAN-GQEIAVK 1758
Query: 471 KISRGSRQGKKEYVTEVKVISQLRHRNLVKLLGWCHDKGEFLLIYEFMPNGSLDSHLFGK 530
++S S QG +E+ E+K+I++L+HRNLVKL G + E N + L
Sbjct: 1759 RLSNTSGQGMEEFKNEIKLIARLQHRNLVKLFGCSVHQDE-----NSHANKKMKILLDST 1923
Query: 531 RTPL-SWGVRHKITLGLASGLLYLHEEWERCVVHRDIKSSNVMLDSSFNVKLGDFGLAKL 589
R+ L W R +I G+A GLLYLH++ ++HRD+K+SN++LD N K+ DFGLA++
Sbjct: 1924 RSKLVDWNKRLQIIDGIARGLLYLHQDSRLRIIHRDLKTSNILLDDEMNPKISDFGLARI 2103
Query: 590 -MDHELGPQTTGLAGTFGYLAPEYVSTGRASKESDVYSFGIVVLEITSGKKATEVMKEKD 648
+ ++ +T + GT+GY+ PEY G S +SDV+SFG++VLEI SGKK +
Sbjct: 2104 FIGDQVEARTKRVMGTYGYMPPEYAVHGSFSIKSDVFSFGVIVLEIISGKKIGR-FYDPH 2280
Query: 649 EEKGMIEWVWDHYGKGELLVAMDENLRKDFDEKQVECLMIVGLWCAHPDVSLRPSIRQAI 708
++ W + + L +DE L ++ + V L C RP + +
Sbjct: 2281 HHLNLLSHAWRLWIEERPLELVDELLDDPVIPTEILRYIHVALLCVQRRPENRPDMLSIV 2460
Query: 709 QVLNFEVSMPNLPPKRPV--ATYHAP--TPSVSSVEASITNSLQDGR 751
+LN E +P P+ P H P S S SIT SL + R
Sbjct: 2461 LMLNGEKELPK--PRLPAFYTGKHDPIWLGSPSRCSTSITISLLEAR 2595
>AI967315
Length = 1308
Score = 177 bits (449), Expect = 5e-45
Identities = 102/291 (35%), Positives = 163/291 (55%), Gaps = 8/291 (2%)
Frame = +1
Query: 429 FTYKELELATNNFSKDRKLGQGGFGAVFKGYFADLDLQVAVKKISRGSRQGKKE--YVTE 486
F+Y+EL ATN FS + +G+GG+ V+KG D ++AVK+++R R +KE ++TE
Sbjct: 112 FSYEELFHATNGFSSENMVGKGGYAEVYKGRLESGD-EIAVKRLTRTCRDERKEKEFLTE 288
Query: 487 VKVISQLRHRNLVKLLGWCHDKGEFLLIYEFMPNGSLDSHLFG-KRTPLSWGVRHKITLG 545
+ I + H N++ LLG C D G +L ++E GS+ S + K PL W R+KI LG
Sbjct: 289 IGTIGHVCHSNVMPLLGCCIDNGLYL-VFELSTVGSVASLIHDEKMAPLDWKTRYKIVLG 465
Query: 546 LASGLLYLHEEWERCVVHRDIKSSNVMLDSSFNVKLGDFGLAKLM-----DHELGPQTTG 600
A GL YLH+ +R ++HRDIK+SN++L F ++ DFGLAK + H + P
Sbjct: 466 TARGLHYLHKGCQRRIIHRDIKASNILLTEDFEPQISDFGLAKWLPSQWTHHSIAP---- 633
Query: 601 LAGTFGYLAPEYVSTGRASKESDVYSFGIVVLEITSGKKATEVMKEKDEEKGMIEWVWDH 660
+ GTFG+LAPEY G +++DV++FG+ +LE+ SG+K + + + W
Sbjct: 634 IEGTFGHLAPEYYMHGVVDEKTDVFAFGVFLLEVISGRKPVD-----GSHQSLHTWAKPI 798
Query: 661 YGKGELLVAMDENLRKDFDEKQVECLMIVGLWCAHPDVSLRPSIRQAIQVL 711
K E+ +D L +D Q + C + RP++ + ++V+
Sbjct: 799 LSKWEIEKLVDPRLEGCYDVTQFNRVAFAASLCIRASSTWRPTMSEVLEVM 951
>TC12704 UP|CAE45596 (CAE45596) S-receptor kinase-like protein 3, complete
Length = 2481
Score = 170 bits (431), Expect = 7e-43
Identities = 102/267 (38%), Positives = 152/267 (56%), Gaps = 9/267 (3%)
Frame = +1
Query: 382 AVIIGVGALVAYVILKRKRKRSEKQKEEAMHLTSMN--------DDLERGAGPRRFTYKE 433
A II + L + +RK++E++ E + +N +D++ F +
Sbjct: 1261 AFIICITILGLATVTCIRRKKNEREDEGGIETRIINHWKDKRGDEDIDLAT---IFDFST 1431
Query: 434 LELATNNFSKDRKLGQGGFGAVFKGYFADLDLQVAVKKISRGSRQGKKEYVTEVKVISQL 493
+ TN+FS+ KLG+GGFG V+KG A+ ++AVK++S S QG +E+ EVK+I++L
Sbjct: 1432 ISSTTNHFSESNKLGEGGFGPVYKGVLAN-GQEIAVKRLSNTSGQGMEEFKNEVKLIARL 1608
Query: 494 RHRNLVKLLGWCHDKGEFLLIYEFMPNGSLDSHLFGKRTPLSWGVRHKITLGLASGLLYL 553
+HRNLVKLLG E LLIYEFM N SLD +F R
Sbjct: 1609 QHRNLVKLLGCSIHHDEMLLIYEFMHNRSLDYFIFDSRLR-------------------- 1728
Query: 554 HEEWERCVVHRDIKSSNVMLDSSFNVKLGDFGLAKLM-DHELGPQTTGLAGTFGYLAPEY 612
++HRD+K+SN++LDS N K+ DFGLA++ ++ +T + GT+GY++PEY
Sbjct: 1729 -------IIHRDLKTSNILLDSEMNPKISDFGLARIFTGDQVEAKTKRVMGTYGYMSPEY 1887
Query: 613 VSTGRASKESDVYSFGIVVLEITSGKK 639
G S +SDV+SFG++VLEI SGKK
Sbjct: 1888 AVHGSFSVKSDVFSFGVIVLEIISGKK 1968
>BP032158
Length = 555
Score = 168 bits (425), Expect = 3e-42
Identities = 85/181 (46%), Positives = 123/181 (66%), Gaps = 3/181 (1%)
Frame = +2
Query: 429 FTYKELELATNNFSKDRKLGQGGFGAVFKGYFADLDLQVAVKKISRGSRQGKKEYVTEVK 488
F+ ++++ ATNNF K+G+GGFG V+KG ++ D+ +AVK++S S+QG +E++ E+
Sbjct: 14 FSLRQIKAATNNFDPANKIGEGGFGPVYKGVLSEGDV-IAVKQLSSKSKQGNREFINEIG 190
Query: 489 VISQLRHRNLVKLLGWCHDKGEFLLIYEFMPNGSLDSHLFG---KRTPLSWGVRHKITLG 545
+IS L+H NLVKL G C + + LL+YE+M N SL LFG ++ L+W R KI +G
Sbjct: 191 MISALQHPNLVKLYGCCIEGNQLLLVYEYMENNSLARALFGNEEQKLNLNWRTRMKICVG 370
Query: 546 LASGLLYLHEEWERCVVHRDIKSSNVMLDSSFNVKLGDFGLAKLMDHELGPQTTGLAGTF 605
+A GL YLHEE +VHRDIK++NV+LD K+ DFGLAKL + E +T +AGT
Sbjct: 371 IAKGLAYLHEESRLKIVHRDIKATNVLLDKDLKAKISDFGLAKLDEEENTHISTRIAGTI 550
Query: 606 G 606
G
Sbjct: 551 G 553
>TC16203 UP|Q8GRU6 (Q8GRU6) LRR receptor-like kinase (Hypernodulation aberrant
root formation protein), complete
Length = 3308
Score = 166 bits (421), Expect = 1e-41
Identities = 102/349 (29%), Positives = 181/349 (51%), Gaps = 14/349 (4%)
Frame = +2
Query: 377 TVSLGAVIIGVG--------ALVAYVILKRKRKRSEKQKEEAMHLTSMNDDLERGAGPRR 428
T + A++IG+ A+ +V+ KR+ R++ K A +
Sbjct: 2024 TARVRAIVIGIALATAVLLVAVTVHVVRKRRLHRAQAWKLTAFQRLEI------------ 2167
Query: 429 FTYKELELATNNFSKDRKLGQGGFGAVFKGYFADLDLQVAVKK-ISRGSRQGKKEYVTEV 487
+ E ++ +G+GG G V++G + VA+K+ + +GS + + E+
Sbjct: 2168 ----KAEDVVECLKEENIIGKGGAGIVYRGSMPN-GTDVAIKRLVGQGSGRNDYGFRAEI 2332
Query: 488 KVISQLRHRNLVKLLGWCHDKGEFLLIYEFMPNGSLDSHLFG-KRTPLSWGVRHKITLGL 546
+ + ++RHRN+++LLG+ +K LL+YE+MPNGSL L G K L W +R+KI +
Sbjct: 2333 ETLGKIRHRNIMRLLGYVSNKDTNLLLYEYMPNGSLGEWLHGAKGGHLRWEMRYKIAVEA 2512
Query: 547 ASGLLYLHEEWERCVVHRDIKSSNVMLDSSFNVKLGDFGLAK-LMDHELGPQTTGLAGTF 605
A GL Y+H + ++HRD+KS+N++LD+ F + DFGLAK L D + +AG++
Sbjct: 2513 ARGLCYMHHDCSPLIIHRDVKSNNILLDADFEAHVADFGLAKFLYDPGASQSMSSIAGSY 2692
Query: 606 GYLAPEYVSTGRASKESDVYSFGIVVLEITSGKKATEVMKEKDEEKGMIEWVWDHYGKGE 665
GY+APEY T + ++SDVYSFG+V+LE+ G+K + + G + +
Sbjct: 2693 GYIAPEYAYTLKVDEKSDVYSFGVVLLELIIGRKPVGEFGDGVDIVGWVNKTMSELSQPS 2872
Query: 666 ---LLVAMDENLRKDFDEKQVECLMIVGLWCAHPDVSLRPSIRQAIQVL 711
L++A+ + + V + + + C RP++R+ + +L
Sbjct: 2873 DTALVLAVVDPRLSGYPLTSVIHMFNIAMMCVKEMGPARPTMREVVHML 3019
>TC16902 homologue to UP|Q8VZH5 (Q8VZH5) Receptor protein kinase-like
protein, partial (46%)
Length = 941
Score = 166 bits (420), Expect = 1e-41
Identities = 103/286 (36%), Positives = 159/286 (55%), Gaps = 15/286 (5%)
Frame = +3
Query: 447 LGQGGFGAVFKGYFADLDLQVAVKKISRGSRQGKKEYVTEVKVISQLRHRNLVKLLGWCH 506
LG GGFG V+ G D +VA+K+ + S QG E+ TE++++S+LRHR+LV L+G+C
Sbjct: 15 LGVGGFGKVYYGE-VDGGTKVAIKRGNPLSEQGVHEFQTEIEMLSKLRHRHLVSLIGYCE 191
Query: 507 DKGEFLLIYEFMPNGSLDSHLF-GKRTPLSWGVRHKITLGLASGLLYLHEEWERCVVHRD 565
+ E +L+Y+ M G+L HL+ ++ PL W R +I +G A GL YLH + ++HRD
Sbjct: 192 ENTEMILVYDHMAYGTLREHLYKTQKPPLPWKQRLEICIGAARGLHYLHTGAKYTIIHRD 371
Query: 566 IKSSNVMLDSSFNVKLGDFGLAKLMDHELGPQ------TTGLAGTFGYLAPEYVSTGRAS 619
+K++N++LD + K+ DFGL+K GP +T + G+FGYL PEY + +
Sbjct: 372 VKTTNILLDEKWVAKVSDFGLSK-----TGPTLDNTHVSTVVKGSFGYLDPEYFRRQQLT 536
Query: 620 KESDVYSFGIVVLEITSGKKATEVMKEKDEEKGMIEWVWDHYGKGELLVAMDENLR---- 675
+SDVYSFG+V+ EI + A K E+ + EW Y KG L +D L+
Sbjct: 537 DKSDVYSFGVVLFEILCARPALNPSLAK-EQVSLAEWASHCYNKGILDQILDPYLKGKIA 713
Query: 676 ----KDFDEKQVECLMIVGLWCAHPDVSLRPSIRQAIQVLNFEVSM 717
K F E ++C+ G+ RPS+ + L F + +
Sbjct: 714 PECFKKFAETAMKCVSDQGI--------ERPSMGDVLWNLEFALQL 827
>TC14667 homologue to UP|Q84P43 (Q84P43) Protein kinase Pti1, complete
Length = 1591
Score = 162 bits (411), Expect = 1e-40
Identities = 96/300 (32%), Positives = 162/300 (54%), Gaps = 10/300 (3%)
Frame = +2
Query: 433 ELELATNNFSKDRKLGQGGFGAVFKGYFADLDLQVAVKKISRGSR-QGKKEYVTEVKVIS 491
EL+ T+NF +G+G +G V+ D + VAVKK+ S + E++T+V ++S
Sbjct: 335 ELKEKTDNFGSKALIGEGSYGRVYYATLNDGNA-VAVKKLDVSSEPETNNEFLTQVSMVS 511
Query: 492 QLRHRNLVKLLGWCHDKGEFLLIYEFMPNGSLDSHLFGKR--------TPLSWGVRHKIT 543
+L++ N V+L G+C + +L YEF GSL L G++ L W R +I
Sbjct: 512 RLKNDNFVELHGYCVEGNLRVLAYEFATMGSLHDILHGRKGVQGAQPGPTLDWIQRVRIA 691
Query: 544 LGLASGLLYLHEEWERCVVHRDIKSSNVMLDSSFNVKLGDFGLA-KLMDHELGPQTTGLA 602
+ A GL YLHE+ + ++HRDI+SSNV++ + K+ DF L+ + D +T +
Sbjct: 692 VDAARGLEYLHEKVQPAIIHRDIRSSNVLIFEDYKAKIADFNLSNQAPDMAARLHSTRVL 871
Query: 603 GTFGYLAPEYVSTGRASKESDVYSFGIVVLEITSGKKATEVMKEKDEEKGMIEWVWDHYG 662
GTFGY APEY TG+ +++SDVYSFG+V+LE+ +G+K + + ++ ++ W
Sbjct: 872 GTFGYHAPEYAMTGQLTQKSDVYSFGVVLLELLTGRKPVDHTMPRGQQ-SLVTWATPRLS 1048
Query: 663 KGELLVAMDENLRKDFDEKQVECLMIVGLWCAHPDVSLRPSIRQAIQVLNFEVSMPNLPP 722
+ ++ +D L+ ++ K V L V C + RP++ ++ L + P P
Sbjct: 1049EDKVKQCVDPKLKGEYPPKGVAKLAAVAALCVQYEAEFRPNMSIVVKALQPLLKTPAAAP 1228
>TC19546 similar to UP|Q40543 (Q40543) Protein-serine/threonine kinase ,
partial (46%)
Length = 731
Score = 160 bits (405), Expect = 7e-40
Identities = 89/212 (41%), Positives = 135/212 (62%), Gaps = 3/212 (1%)
Frame = +1
Query: 402 RSEK-QKEEAMHLTSM--NDDLERGAGPRRFTYKELELATNNFSKDRKLGQGGFGAVFKG 458
RSE QK+ A S ND +G +++YK+++ AT NF+ LGQG FG V+K
Sbjct: 109 RSENLQKKSAFSWWSHQNNDQFASPSGIPKYSYKDIQKATQNFTTI--LGQGSFGTVYKA 282
Query: 459 YFADLDLQVAVKKISRGSRQGKKEYVTEVKVISQLRHRNLVKLLGWCHDKGEFLLIYEFM 518
++ VAVK ++ S+QG+ E+ TEV ++ +L HRNLV L+G+C DKG+ +L+Y+FM
Sbjct: 283 TMPTGEV-VAVKVLAPNSKQGEHEFQTEVHLLGRLHHRNLVNLVGFCVDKGQRILVYQFM 459
Query: 519 PNGSLDSHLFGKRTPLSWGVRHKITLGLASGLLYLHEEWERCVVHRDIKSSNVMLDSSFN 578
NGSL + L+G+ LSW R +I + ++ G+ YLHE V+HRD+KS+N++LD S
Sbjct: 460 SNGSLANLLYGEEKELSWDERLQIAMDISHGIEYLHEGAVPPVIHRDLKSANILLDDSMR 639
Query: 579 VKLGDFGLAKLMDHELGPQTTGLAGTFGYLAP 610
+ DFGL+K + + +GL GT+GY+ P
Sbjct: 640 AMVADFGLSK--EEIFDGRNSGLKGTYGYMDP 729
>AV422094
Length = 462
Score = 158 bits (400), Expect = 3e-39
Identities = 77/149 (51%), Positives = 106/149 (70%)
Frame = +2
Query: 426 PRRFTYKELELATNNFSKDRKLGQGGFGAVFKGYFADLDLQVAVKKISRGSRQGKKEYVT 485
P F+Y EL+ ATN+F+ D KLG+GGFG V+KG D + +AVK++S GS QGK +++
Sbjct: 5 PYTFSYSELKNATNDFNIDNKLGEGGFGPVYKGILNDGTV-IAVKQLSLGSHQGKSQFIA 181
Query: 486 EVKVISQLRHRNLVKLLGWCHDKGEFLLIYEFMPNGSLDSHLFGKRTPLSWGVRHKITLG 545
E+ IS ++HRNLVKL G C + + LL+YE++ N SLD LFGK L+W R+ I LG
Sbjct: 182 EIATISAVQHRNLVKLYGCCIEGSKRLLVYEYLENKSLDQALFGKALSLNWSTRYDICLG 361
Query: 546 LASGLLYLHEEWERCVVHRDIKSSNVMLD 574
+A GL YLHEE +VHRD+K+SN++LD
Sbjct: 362 VARGLTYLHEESRLRIVHRDVKASNILLD 448
>TC12343 similar to UP|CDK9_SCHPO (Q96WV9) Serine/threonine protein kinase
Cdk9 (Cyclin-dependent kinase Cdk9) , partial (7%)
Length = 723
Score = 158 bits (399), Expect = 3e-39
Identities = 80/180 (44%), Positives = 118/180 (65%), Gaps = 4/180 (2%)
Frame = +3
Query: 418 DDLERGAGP--RRFTYKELELATNNFSKDRKLGQGGFGAVFKGYFADLDLQVAVKKISRG 475
DD++ A + F+Y+ L AT NF KLG+GGFG VFKG D ++AVKK+SR
Sbjct: 168 DDIQNIAAKEHKTFSYETLVAATKNFHAVNKLGEGGFGPVFKGKLND-GREIAVKKLSRR 344
Query: 476 SRQGKKEYVTEVKVISQLRHRNLVKLLGWCHDKGEFLLIYEFMPNGSLDSHLF--GKRTP 533
S QG+ +++ E K++++++HRN+V L G+C E LL+YE++P SLD LF K+
Sbjct: 345 SNQGRTQFINEAKLLTRVQHRNVVSLFGYCAHGSEKLLVYEYVPRESLDKLLFRSQKKEQ 524
Query: 534 LSWGVRHKITLGLASGLLYLHEEWERCVVHRDIKSSNVMLDSSFNVKLGDFGLAKLMDHE 593
L W R I G+A GLLYLHE+ C++HRDIK++N++LD + K+ DFGLA++ +
Sbjct: 525 LDWKRRFDIISGVARGLLYLHEDSHDCIIHRDIKAANILLDEKWVPKIADFGLARIFPED 704
>TC8971 similar to GB|AAM16251.1|20334780|AY093990 At2g11520/F14P14.15
{Arabidopsis thaliana;}, partial (37%)
Length = 1079
Score = 153 bits (386), Expect = 1e-37
Identities = 87/224 (38%), Positives = 138/224 (60%), Gaps = 4/224 (1%)
Frame = +3
Query: 486 EVKVISQLRHRNLVKLLGWCHDKGEFLLIYEFMPNGSLDSHLFGKRTP-LSWGVRHKITL 544
EV++++++ HRNLVKLLG+ E +LI E++PNG+L HL G R L + R +I +
Sbjct: 6 EVELLAKIDHRNLVKLLGFIDKGNERILITEYVPNGTLREHLDGLRGKILDFNQRLEIAI 185
Query: 545 GLASGLLYLHEEWERCVVHRDIKSSNVMLDSSFNVKLGDFGLAKLMDHELGPQT---TGL 601
+A GL YLH E+ ++HRD+KSSN++L S K+ DFG A+L G QT T +
Sbjct: 186 DVAHGLTYLHLYAEKQIIHRDVKSSNILLTESMRAKVADFGFARLGPVN-GDQTHISTKV 362
Query: 602 AGTFGYLAPEYVSTGRASKESDVYSFGIVVLEITSGKKATEVMKEKDEEKGMIEWVWDHY 661
GT GYL PEY+ T + + +SDVYSFGI++LEI +G++ E+ K D E+ + W + Y
Sbjct: 363 KGTVGYLDPEYMKTHQLTPKSDVYSFGILLLEILTGRRPVELKKAAD-ERVTLRWAFRKY 539
Query: 662 GKGELLVAMDENLRKDFDEKQVECLMIVGLWCAHPDVSLRPSIR 705
+G ++ MD + + + + ++ + CA P + RP+++
Sbjct: 540 NEGSVVELMDPLMEEAVNADVLMKMLDLSFQCAAPIRTDRPNMK 671
>TC12473 similar to UP|Q9M068 (Q9M068) Protein kinase-like protein, partial
(42%)
Length = 946
Score = 152 bits (383), Expect = 2e-37
Identities = 77/170 (45%), Positives = 111/170 (65%), Gaps = 10/170 (5%)
Frame = +3
Query: 429 FTYKELELATNNFSKDRKLGQGGFGAVFKGYFADLDLQ---------VAVKKISRGSRQG 479
F++ +L+ AT +F D +G+GGFG V+KG+ + L VAVKK++ S QG
Sbjct: 420 FSFGDLKSATKSFKADALIGEGGFGKVYKGWLDEKKLSPTKPGSGIMVAVKKLNPESMQG 599
Query: 480 KKEYVTEVKVISQLRHRNLVKLLGWCHDKGEFLLIYEFMPNGSLDSHLFGKRT-PLSWGV 538
E+ +E+ + ++ H NLVKLLG+C D EFLL+YEFMP GSL++HLF + T LSW
Sbjct: 600 FHEWQSEINFLGRISHPNLVKLLGYCRDDEEFLLVYEFMPRGSLENHLFRRNTESLSWNT 779
Query: 539 RHKITLGLASGLLYLHEEWERCVVHRDIKSSNVMLDSSFNVKLGDFGLAK 588
R KI +G A GL +LH + V++RD K+SN++LD ++N K+ FGL K
Sbjct: 780 RLKIAIGAARGLAFLH-SLXKIVIYRDFKASNILLDGNYNAKISXFGLTK 926
>TC9538 similar to UP|RLK5_ARATH (P47735) Receptor-like protein kinase 5
precursor , partial (10%)
Length = 1067
Score = 146 bits (368), Expect = 1e-35
Identities = 88/233 (37%), Positives = 132/233 (55%), Gaps = 18/233 (7%)
Frame = +2
Query: 497 NLVKLLGWCHDKGEFLLIYEFMPNGSLDSHLFGKR------------TPLSWGVRHKITL 544
N+V+LL ++ LL+YE++ N SLD L K T L W R KI +
Sbjct: 5 NIVRLLCCISNEASMLLVYEYLENHSLDKWLHLKPKSSSVSGVVQQYTVLDWPKRLKIAI 184
Query: 545 GLASGLLYLHEEWERCVVHRDIKSSNVMLDSSFNVKLGDFGLAKLM--DHELGPQTTGLA 602
G A GL Y+H + +VHRD+K+SN++LD FN K+ DFGLA+++ EL +T +
Sbjct: 185 GAAQGLSYMHHDCSPPIVHRDVKTSNILLDKQFNAKVADFGLARMLIKPGELNIMST-VI 361
Query: 603 GTFGYLAPEYVSTGRASKESDVYSFGIVVLEITSGKKATEVMKEKDEEKGMIEWVWDHYG 662
GTFGY+APEYV T R S++ DVYSFG+V+LE+T+GK+A D+ + EW W H
Sbjct: 362 GTFGYIAPEYVQTTRISEKVDVYSFGVVLLELTTGKEA----NYGDQHSSLAEWAWRHIL 529
Query: 663 KGELLVAMDENLRKDFDE----KQVECLMIVGLWCAHPDVSLRPSIRQAIQVL 711
G + + L KD E ++ + +G+ C + RPS+++ + +L
Sbjct: 530 IGS---NVXDLLXKDVMEASYIDEMCSVFKLGVMCTATLPATRPSMKEVLPIL 679
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.319 0.136 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,470,688
Number of Sequences: 28460
Number of extensions: 203569
Number of successful extensions: 1797
Number of sequences better than 10.0: 314
Number of HSP's better than 10.0 without gapping: 1590
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1608
length of query: 751
length of database: 4,897,600
effective HSP length: 97
effective length of query: 654
effective length of database: 2,136,980
effective search space: 1397584920
effective search space used: 1397584920
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 59 (27.3 bits)
Medicago: description of AC146709.2


