

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146709.1 - phase: 0 /pseudo
(495 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
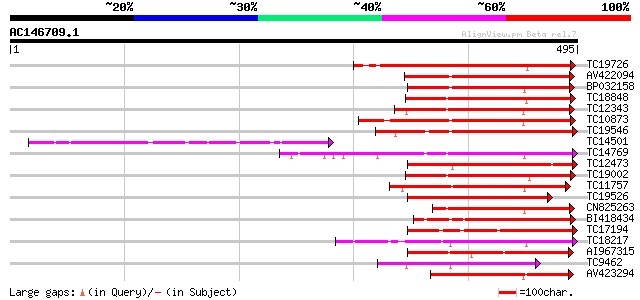
Score E
Sequences producing significant alignments: (bits) Value
TC19726 similar to UP|Q9XIS3 (Q9XIS3) Lectin-like protein kinase... 179 1e-45
AV422094 160 6e-40
BP032158 145 1e-35
TC18848 weakly similar to UP|Q9SSQ6 (Q9SSQ6) F6D8.24 protein, pa... 145 2e-35
TC12343 similar to UP|CDK9_SCHPO (Q96WV9) Serine/threonine prote... 144 4e-35
TC10873 similar to UP|Q9LDZ5 (Q9LDZ5) F2D10.13 (F5M15.3), partia... 143 7e-35
TC19546 similar to UP|Q40543 (Q40543) Protein-serine/threonine k... 142 1e-34
TC14501 similar to UP|LEC_LOTTE (P19664) Anti-H(O) lectin (LTA),... 142 1e-34
TC14769 UP|Q8LKX1 (Q8LKX1) Receptor-like kinase SYMRK, complete 141 3e-34
TC12473 similar to UP|Q9M068 (Q9M068) Protein kinase-like protei... 140 4e-34
TC19002 weakly similar to UP|Q9XJ10 (Q9XJ10) ESTs C22458(C62866)... 140 5e-34
TC11757 UP|Q91W27 (Q91W27) Tuberoinfundibular 39 residue protein... 135 2e-32
TC19526 homologue to UP|AAQ96340 (AAQ96340) Protein kinase-like ... 134 4e-32
CN825263 131 2e-31
BI418434 125 1e-29
TC17194 UP|CAE02590 (CAE02590) Nod-factor receptor 1b, complete 125 2e-29
TC18217 weakly similar to UP|Q9FLJ8 (Q9FLJ8) Receptor protein ki... 120 7e-28
AI967315 117 4e-27
TC9462 similar to UP|O49840 (O49840) Protein kinase (AT2G02800/T... 117 6e-27
AV423294 117 6e-27
>TC19726 similar to UP|Q9XIS3 (Q9XIS3) Lectin-like protein kinase, partial
(30%)
Length = 728
Score = 179 bits (453), Expect = 1e-45
Identities = 94/199 (47%), Positives = 130/199 (65%), Gaps = 5/199 (2%)
Frame = +3
Query: 301 VIVIGVGALVAYALFWRKRKRSNKQKEEAMHLTSMNDDLERGAGPRRFTYKELDLATNNF 360
VIV +G L L+W + K K++EE + + PR F Y EL ATNNF
Sbjct: 12 VIVAVIGGL----LWWIRLK---KKREEGSNSQILGTLKSLPGTPREFNYVELKKATNNF 170
Query: 361 SKDRKLGQGGFGAVYKGYFADLDLQVAVKKISRGSRQGKKEYVTEVKVISQLRHRNLVKL 420
+ KLGQGG+G VY+G L+VAVK SR + ++++E+ +I++LRH++LV+L
Sbjct: 171 DEKHKLGQGGYGVVYRGMLPKEKLEVAVKMFSRDKMKSTDDFLSELIIINRLRHKHLVRL 350
Query: 421 LGWCHDKGEFLLVYEFMPNGSLDSHLFGKR-----TPLSWGVRHKIALGLASGLLYLHEE 475
GWCH G LLVY++MPNGSLDSH+F + TPLSW +R+KI G+AS L YLH E
Sbjct: 351 QGWCHKNGVLLLVYDYMPNGSLDSHIFCEEGGTITTPLSWPLRYKIISGVASALNYLHNE 530
Query: 476 WERCVVHRDIKSSNVMLDS 494
+++ VVHRD+K+SN+MLDS
Sbjct: 531 YDQKVVHRDLKASNIMLDS 587
>AV422094
Length = 462
Score = 160 bits (404), Expect = 6e-40
Identities = 79/149 (53%), Positives = 105/149 (70%)
Frame = +2
Query: 345 PRRFTYKELDLATNNFSKDRKLGQGGFGAVYKGYFADLDLQVAVKKISRGSRQGKKEYVT 404
P F+Y EL ATN+F+ D KLG+GGFG VYKG D + +AVK++S GS QGK +++
Sbjct: 5 PYTFSYSELKNATNDFNIDNKLGEGGFGPVYKGILNDGTV-IAVKQLSLGSHQGKSQFIA 181
Query: 405 EVKVISQLRHRNLVKLLGWCHDKGEFLLVYEFMPNGSLDSHLFGKRTPLSWGVRHKIALG 464
E+ IS ++HRNLVKL G C + + LLVYE++ N SLD LFGK L+W R+ I LG
Sbjct: 182 EIATISAVQHRNLVKLYGCCIEGSKRLLVYEYLENKSLDQALFGKALSLNWSTRYDICLG 361
Query: 465 LASGLLYLHEEWERCVVHRDIKSSNVMLD 493
+A GL YLHEE +VHRD+K+SN++LD
Sbjct: 362 VARGLTYLHEESRLRIVHRDVKASNILLD 448
>BP032158
Length = 555
Score = 145 bits (366), Expect = 1e-35
Identities = 73/149 (48%), Positives = 104/149 (68%), Gaps = 3/149 (2%)
Frame = +2
Query: 348 FTYKELDLATNNFSKDRKLGQGGFGAVYKGYFADLDLQVAVKKISRGSRQGKKEYVTEVK 407
F+ +++ ATNNF K+G+GGFG VYKG ++ D+ +AVK++S S+QG +E++ E+
Sbjct: 14 FSLRQIKAATNNFDPANKIGEGGFGPVYKGVLSEGDV-IAVKQLSSKSKQGNREFINEIG 190
Query: 408 VISQLRHRNLVKLLGWCHDKGEFLLVYEFMPNGSLDSHLFG---KRTPLSWGVRHKIALG 464
+IS L+H NLVKL G C + + LLVYE+M N SL LFG ++ L+W R KI +G
Sbjct: 191 MISALQHPNLVKLYGCCIEGNQLLLVYEYMENNSLARALFGNEEQKLNLNWRTRMKICVG 370
Query: 465 LASGLLYLHEEWERCVVHRDIKSSNVMLD 493
+A GL YLHEE +VHRDIK++NV+LD
Sbjct: 371 IAKGLAYLHEESRLKIVHRDIKATNVLLD 457
>TC18848 weakly similar to UP|Q9SSQ6 (Q9SSQ6) F6D8.24 protein, partial (64%)
Length = 969
Score = 145 bits (365), Expect = 2e-35
Identities = 76/152 (50%), Positives = 105/152 (69%), Gaps = 3/152 (1%)
Frame = +2
Query: 346 RRFTYKELDLATNNFSKDRKLGQGGFGAVYKGYFADLDLQVAVKKISRGSRQGKKEYVTE 405
R FTYKEL AT FS D KLG+GGFG+VY G +D LQ+AVKK+ + + + E+ E
Sbjct: 260 RIFTYKELHAATGGFSDDNKLGEGGFGSVYWGRTSD-GLQIAVKKLKAMNSKAEMEFAVE 436
Query: 406 VKVISQLRHRNLVKLLGWCHDKGEFLLVYEFMPNGSLDSHLFGK---RTPLSWGVRHKIA 462
V+V+ ++RH+NL+ L G+C + L+VY++MPN SL SHL G+ L+W R KIA
Sbjct: 437 VEVLGRVRHKNLLGLRGYCVGDDQRLIVYDYMPNLSLLSHLHGQFAVEVQLNWQKRMKIA 616
Query: 463 LGLASGLLYLHEEWERCVVHRDIKSSNVMLDS 494
+G A G+LYLH E ++HRDIK+SNV+L+S
Sbjct: 617 IGSAEGILYLHHEVTPHIIHRDIKASNVLLNS 712
>TC12343 similar to UP|CDK9_SCHPO (Q96WV9) Serine/threonine protein kinase
Cdk9 (Cyclin-dependent kinase Cdk9) , partial (7%)
Length = 723
Score = 144 bits (362), Expect = 4e-35
Identities = 74/161 (45%), Positives = 107/161 (65%), Gaps = 4/161 (2%)
Frame = +3
Query: 337 DDLERGAGP--RRFTYKELDLATNNFSKDRKLGQGGFGAVYKGYFADLDLQVAVKKISRG 394
DD++ A + F+Y+ L AT NF KLG+GGFG V+KG D ++AVKK+SR
Sbjct: 168 DDIQNIAAKEHKTFSYETLVAATKNFHAVNKLGEGGFGPVFKGKLND-GREIAVKKLSRR 344
Query: 395 SRQGKKEYVTEVKVISQLRHRNLVKLLGWCHDKGEFLLVYEFMPNGSLDSHLF--GKRTP 452
S QG+ +++ E K++++++HRN+V L G+C E LLVYE++P SLD LF K+
Sbjct: 345 SNQGRTQFINEAKLLTRVQHRNVVSLFGYCAHGSEKLLVYEYVPRESLDKLLFRSQKKEQ 524
Query: 453 LSWGVRHKIALGLASGLLYLHEEWERCVVHRDIKSSNVMLD 493
L W R I G+A GLLYLHE+ C++HRDIK++N++LD
Sbjct: 525 LDWKRRFDIISGVARGLLYLHEDSHDCIIHRDIKAANILLD 647
>TC10873 similar to UP|Q9LDZ5 (Q9LDZ5) F2D10.13 (F5M15.3), partial (77%)
Length = 1478
Score = 143 bits (360), Expect = 7e-35
Identities = 83/193 (43%), Positives = 116/193 (60%), Gaps = 3/193 (1%)
Frame = +1
Query: 305 GVGALVAYALFWRKRKRSNKQKEEAMHLTSMNDDLERGAGPRRFTYKELDLATNNFSKDR 364
GV A A RKR+ K K ++S N F ++EL AT NF +
Sbjct: 298 GVNAAAAATDGERKRETKGKGKG----VSSSNGSSNGKTAAASFGFRELADATRNFKEAN 465
Query: 365 KLGQGGFGAVYKGYFADLDLQVAVKKISRGSRQGKKEYVTEVKVISQLRHRNLVKLLGWC 424
+G+GGFG VYKG + VAVK++S RQG +E+V EV ++S L H NLV+L+G+C
Sbjct: 466 LIGEGGFGKVYKGRLTTGEA-VAVKQLSHDGRQGFQEFVMEVLMLSLLHHTNLVRLIGYC 642
Query: 425 HDKGEFLLVYEFMPNGSLDSHLF---GKRTPLSWGVRHKIALGLASGLLYLHEEWERCVV 481
D + LLVYE+MP GSL+ HLF + PL+W R K+A+G A GL YLH + V+
Sbjct: 643 TDGDQRLLVYEYMPMGSLEDHLFELSHDKEPLNWSTRMKVAVGAARGLEYLHCTADPPVI 822
Query: 482 HRDIKSSNVMLDS 494
+RD+KS+N++LD+
Sbjct: 823 YRDLKSANILLDN 861
>TC19546 similar to UP|Q40543 (Q40543) Protein-serine/threonine kinase ,
partial (46%)
Length = 731
Score = 142 bits (359), Expect = 1e-34
Identities = 78/178 (43%), Positives = 115/178 (63%), Gaps = 2/178 (1%)
Frame = +1
Query: 320 KRSNKQKEEAMHLTSM--NDDLERGAGPRRFTYKELDLATNNFSKDRKLGQGGFGAVYKG 377
+ N QK+ A S ND +G +++YK++ AT NF+ LGQG FG VYK
Sbjct: 109 RSENLQKKSAFSWWSHQNNDQFASPSGIPKYSYKDIQKATQNFTTI--LGQGSFGTVYKA 282
Query: 378 YFADLDLQVAVKKISRGSRQGKKEYVTEVKVISQLRHRNLVKLLGWCHDKGEFLLVYEFM 437
++ VAVK ++ S+QG+ E+ TEV ++ +L HRNLV L+G+C DKG+ +LVY+FM
Sbjct: 283 TMPTGEV-VAVKVLAPNSKQGEHEFQTEVHLLGRLHHRNLVNLVGFCVDKGQRILVYQFM 459
Query: 438 PNGSLDSHLFGKRTPLSWGVRHKIALGLASGLLYLHEEWERCVVHRDIKSSNVMLDSS 495
NGSL + L+G+ LSW R +IA+ ++ G+ YLHE V+HRD+KS+N++LD S
Sbjct: 460 SNGSLANLLYGEEKELSWDERLQIAMDISHGIEYLHEGAVPPVIHRDLKSANILLDDS 633
>TC14501 similar to UP|LEC_LOTTE (P19664) Anti-H(O) lectin (LTA), partial
(86%)
Length = 1066
Score = 142 bits (358), Expect = 1e-34
Identities = 99/269 (36%), Positives = 150/269 (54%), Gaps = 3/269 (1%)
Frame = +1
Query: 17 TQSMASSLYILTLFYMSLIFFNIPASSIHFKYPSFDPSDANIVYQGSAAP-RDGEVNFNI 75
+ + + + LF++ I N+ A++I F + F D +++ QG A DG +
Sbjct: 58 SNASTQNFLLACLFFLLAINVNL-AAAISFNFTEFT-DDGSLIIQGDAKIWADGSLALPT 231
Query: 76 NENYSCQVGRVFYSEKVLLWDSNTGKLTDFTTHYTFVI-NTQGRSPSLYGHGLAFFLVPY 134
+ + R Y+ V +WDS TG + F T ++F+I + + +P+ GL FFL P+
Sbjct: 232 DPSVGFTTSRALYATPVPIWDSTTGNVASFVTSFSFIIKDFEDYNPA---DGLVFFLAPF 402
Query: 135 GFEIPLNSDGGFMGLFNTTTMVSSSNQIVHVEFDSFANREFREARGHVGININSIISSAT 194
G EIP S GG G+ N + NQIV VEFD+F N + + H+GI++NS+IS T
Sbjct: 403 GTEIPKESTGGRFGIINGK---DAFNQIVAVEFDTFIN-PWDSSPRHIGIDVNSLISLKT 570
Query: 195 TPWNASKHSGDTAEVWIRYNSTTKNLTVSWKYQTTSNPQENTSLSISIDLMKVMPEWITV 254
PWN K +G +V I Y+S TK L+V ++ N Q +T +S IDL V+PE ++V
Sbjct: 571 VPWN--KVAGSLEKVTIIYDSQTKTLSVLVIHE---NGQIST-ISQEIDLKVVLPEEVSV 732
Query: 255 GFSA-ATSYVQELNYLLSWEFNSTLATSG 282
GFSA TS +E + + SW F STL T+G
Sbjct: 733 GFSATTTSGGRERHDIYSWSFTSTLNTNG 819
>TC14769 UP|Q8LKX1 (Q8LKX1) Receptor-like kinase SYMRK, complete
Length = 3495
Score = 141 bits (355), Expect = 3e-34
Identities = 104/286 (36%), Positives = 154/286 (53%), Gaps = 26/286 (9%)
Frame = +3
Query: 236 TSLSISIDL-----MKVMPEWITVGFSAATSYVQELNYLLSWE----FNSTLATS--GDS 284
+SL IS+DL M +PE I V S N +S E NS+L + G
Sbjct: 1731 SSLLISVDLSYNDLMGKLPESI-VKLPHLKSLYFGCNEHMSPEDPANMNSSLINTDYGRC 1907
Query: 285 KTKETR---LVIILTVSCGVIVIGVGALVAYALFWRKR---------KRSNKQKEEAMHL 332
K KE+R +++I ++CG ++I + V + +R++ K+ + L
Sbjct: 1908 KGKESRFGQVIVIGAITCGSLLITLAFGVLFVCRYRQKLIPWEGFAGKKYPMETNIIFSL 2087
Query: 333 TSMNDDLERGAGPRRFTYKELDLATNNFSKDRKLGQGGFGAVYKGYFADLDLQVAVKKIS 392
S +D + + FT + +++AT + +G+GGFG+VY+G D +VAVK S
Sbjct: 2088 PSKDDFFIKSVSIQAFTLEYIEVATERYKT--LIGEGGFGSVYRGTLND-GQEVAVKVRS 2258
Query: 393 RGSRQGKKEYVTEVKVISQLRHRNLVKLLGWCHDKGEFLLVYEFMPNGSLDSHLFG---K 449
S QG +E+ E+ ++S ++H NLV LLG+C++ + +LVY FM NGSL L+G K
Sbjct: 2259 STSTQGTREFDNELNLLSAIQHENLVPLLGYCNESDQQILVYPFMSNGSLQDRLYGEPAK 2438
Query: 450 RTPLSWGVRHKIALGLASGLLYLHEEWERCVVHRDIKSSNVMLDSS 495
R L W R IALG A GL YLH R V+HRDIKSSN++LD S
Sbjct: 2439 RKILDWPTRLSIALGAARGLAYLHTFPGRSVIHRDIKSSNILLDHS 2576
>TC12473 similar to UP|Q9M068 (Q9M068) Protein kinase-like protein, partial
(42%)
Length = 946
Score = 140 bits (354), Expect = 4e-34
Identities = 74/158 (46%), Positives = 103/158 (64%), Gaps = 10/158 (6%)
Frame = +3
Query: 348 FTYKELDLATNNFSKDRKLGQGGFGAVYKGYFADLDLQ---------VAVKKISRGSRQG 398
F++ +L AT +F D +G+GGFG VYKG+ + L VAVKK++ S QG
Sbjct: 420 FSFGDLKSATKSFKADALIGEGGFGKVYKGWLDEKKLSPTKPGSGIMVAVKKLNPESMQG 599
Query: 399 KKEYVTEVKVISQLRHRNLVKLLGWCHDKGEFLLVYEFMPNGSLDSHLFGKRT-PLSWGV 457
E+ +E+ + ++ H NLVKLLG+C D EFLLVYEFMP GSL++HLF + T LSW
Sbjct: 600 FHEWQSEINFLGRISHPNLVKLLGYCRDDEEFLLVYEFMPRGSLENHLFRRNTESLSWNT 779
Query: 458 RHKIALGLASGLLYLHEEWERCVVHRDIKSSNVMLDSS 495
R KIA+G A GL +LH + V++RD K+SN++LD +
Sbjct: 780 RLKIAIGAARGLAFLH-SLXKIVIYRDFKASNILLDGN 890
>TC19002 weakly similar to UP|Q9XJ10 (Q9XJ10) ESTs C22458(C62866), partial
(48%)
Length = 780
Score = 140 bits (353), Expect = 5e-34
Identities = 72/152 (47%), Positives = 104/152 (68%), Gaps = 3/152 (1%)
Frame = +1
Query: 346 RRFTYKELDLATNNFSKDRKLGQGGFGAVYKGYFADLDLQVAVKKISRGSRQGKKEYVTE 405
R F+ KEL ATNNF+ D KLG+GGFG+VY G D Q+AVK++ S + E+ E
Sbjct: 22 RVFSLKELHSATNNFNYDNKLGEGGFGSVYWGQLWD-GSQIAVKRLKVWSNKADMEFAVE 198
Query: 406 VKVISQLRHRNLVKLLGWCHDKGEFLLVYEFMPNGSLDSHLFGKRTP---LSWGVRHKIA 462
V++++++RH+NL+ L G+C + E L+VY++MPN SL SHL G+ + L W R IA
Sbjct: 199 VEILARVRHKNLLSLRGYCAEGQERLIVYDYMPNLSLLSHLHGQHSSECLLDWNRRMNIA 378
Query: 463 LGLASGLLYLHEEWERCVVHRDIKSSNVMLDS 494
+G A G++YLH + ++HRDIK+SNV+LDS
Sbjct: 379 IGSAEGIVYLHHQATPHIIHRDIKASNVLLDS 474
>TC11757 UP|Q91W27 (Q91W27) Tuberoinfundibular 39 residue protein, partial
(10%)
Length = 715
Score = 135 bits (340), Expect = 2e-32
Identities = 75/164 (45%), Positives = 101/164 (60%), Gaps = 6/164 (3%)
Frame = +2
Query: 332 LTSMNDDLER---GAGPRRFTYKELDLATNNFSKDRKLGQGGFGAVYKGYFADLDLQVAV 388
L S N D+++ G R FTYKEL +AT+NFS K+G+GGFG VYKG D + A+
Sbjct: 227 LASTNPDIDKEISGIHVRVFTYKELKIATDNFSPVNKIGEGGFGPVYKGVLKDGKVG-AI 403
Query: 389 KKISRGSRQGKKEYVTEVKVISQLRHRNLVKLLGWCHDKGEFLLVYEFMPNGSLDSHLFG 448
K +S S+QG KE++TE+ VIS++ H NLV+L G C + +LVY + N SL L G
Sbjct: 404 KVLSAESKQGVKEFMTEINVISEIEHENLVQLYGCCVEGNHRILVYNYHENNSLAQTLLG 583
Query: 449 ---KRTPLSWGVRHKIALGLASGLLYLHEEWERCVVHRDIKSSN 489
W R +I +G+A GL +LHEE +VHRDIK+SN
Sbjct: 584 GGHSNIHFDWRTRSRICIGVARGLSFLHEEVRPHIVHRDIKASN 715
>TC19526 homologue to UP|AAQ96340 (AAQ96340) Protein kinase-like protein,
partial (37%)
Length = 432
Score = 134 bits (336), Expect = 4e-32
Identities = 66/127 (51%), Positives = 83/127 (64%)
Frame = +1
Query: 348 FTYKELDLATNNFSKDRKLGQGGFGAVYKGYFADLDLQVAVKKISRGSRQGKKEYVTEVK 407
FT++EL AT NF + LG+GGFG VYKG VAVK++ R QG +E++ EV
Sbjct: 52 FTFRELAAATKNFRPECLLGEGGFGRVYKGRLESTGQVVAVKQLDRNGLQGNREFLVEVL 231
Query: 408 VISQLRHRNLVKLLGWCHDKGEFLLVYEFMPNGSLDSHLFGKRTPLSWGVRHKIALGLAS 467
++S L H NLV L+G+C D + LLVYEFMP GSL+ HL G + PL W R KIA G A
Sbjct: 232 MLSLLHHPNLVNLIGYCADGDQRLLVYEFMPLGSLEDHLHGDKEPLDWNTRMKIAAGAAK 411
Query: 468 GLLYLHE 474
GL YLH+
Sbjct: 412 GLEYLHD 432
>CN825263
Length = 663
Score = 131 bits (330), Expect = 2e-31
Identities = 68/127 (53%), Positives = 89/127 (69%), Gaps = 3/127 (2%)
Frame = +1
Query: 370 GFGAVYKGYFADLDLQVAVKKISRGSRQGKKEYVTEVKVISQLRHRNLVKLLGWCHDKGE 429
GFG VYKG D VAVK + R ++G +E++ EV+++S+L HRNLVKL+G C +K
Sbjct: 1 GFGLVYKGILND-GRDVAVKILKRDDQRGGREFLAEVEMLSRLHHRNLVKLIGICIEKQT 177
Query: 430 FLLVYEFMPNGSLDSHLFG--KRT-PLSWGVRHKIALGLASGLLYLHEEWERCVVHRDIK 486
L+YE +PNGS++SHL G K T PL W R KIALG A GL YLHE+ CV+HRD K
Sbjct: 178 RCLIYELVPNGSVESHLHGADKETGPLDWNARMKIALGAARGLAYLHEDSNPCVIHRDFK 357
Query: 487 SSNVMLD 493
SSN++L+
Sbjct: 358 SSNILLE 378
>BI418434
Length = 478
Score = 125 bits (315), Expect = 1e-29
Identities = 67/142 (47%), Positives = 93/142 (65%)
Frame = +2
Query: 353 LDLATNNFSKDRKLGQGGFGAVYKGYFADLDLQVAVKKISRGSRQGKKEYVTEVKVISQL 412
L LAT FS+ K+G GGFG V++G +D L VAVK++ R G+KE+ EV I +
Sbjct: 2 LQLATRGFSE--KVGHGGFGTVFQGELSDSSL-VAVKRLERPGG-GEKEFRAEVSTIGNI 169
Query: 413 RHRNLVKLLGWCHDKGEFLLVYEFMPNGSLDSHLFGKRTPLSWGVRHKIALGLASGLLYL 472
+H NLV+L G+C + LLVYE+M NG+L ++L LSW VR ++A+G A G+ YL
Sbjct: 170 QHVNLVRLRGFCSENSHRLLVYEYMHNGALSAYLRKDGPCLSWDVRFQVAIGTAKGIAYL 349
Query: 473 HEEWERCVVHRDIKSSNVMLDS 494
HEE C++H DIK N++LDS
Sbjct: 350 HEELRCCILHCDIKPENILLDS 415
>TC17194 UP|CAE02590 (CAE02590) Nod-factor receptor 1b, complete
Length = 2193
Score = 125 bits (313), Expect = 2e-29
Identities = 68/150 (45%), Positives = 99/150 (65%), Gaps = 2/150 (1%)
Frame = +1
Query: 348 FTYKELDLATNNFSKDRKLGQGGFGAVYKGYFADL-DLQVAVKKISRGSRQGKKEYVTEV 406
F+Y+EL ATNNFS D K+GQGGFGAVY +A+L + A+KK+ Q E++ E+
Sbjct: 1051 FSYQELAKATNNFSLDNKIGQGGFGAVY---YAELRGKKTAIKKMDV---QASTEFLCEL 1212
Query: 407 KVISQLRHRNLVKLLGWCHDKGEFLLVYEFMPNGSLDSHLFGK-RTPLSWGVRHKIALGL 465
KV++ + H NLV+L+G+C + G LVYE + NG+L +L G + PL W R +IAL
Sbjct: 1213 KVLTHVHHLNLVRLIGYCVE-GSLFLVYEHIDNGNLGQYLHGSGKEPLPWSSRVQIALDA 1389
Query: 466 ASGLLYLHEEWERCVVHRDIKSSNVMLDSS 495
A GL Y+HE +HRD+KS+N+++D +
Sbjct: 1390 ARGLEYIHEHTVPVYIHRDVKSANILIDKN 1479
>TC18217 weakly similar to UP|Q9FLJ8 (Q9FLJ8) Receptor protein kinase-like
protein, partial (15%)
Length = 688
Score = 120 bits (300), Expect = 7e-28
Identities = 79/218 (36%), Positives = 122/218 (55%), Gaps = 7/218 (3%)
Frame = +3
Query: 285 KTKETRLVIILTVSCGVIVIGVGALVAYALFWRKRKRSNKQKEEAMHLTSMNDDLERGAG 344
K ++ +L+++LT+ G V G + L +K+K + + E ++ ++L
Sbjct: 60 KHQQQQLLLLLTLL*G-FVSQSGRMFLKCLGLKKKKHTPCETERPY--PTVIEEL----- 215
Query: 345 PRRFTYKELDLATNNFSKDRKLGQGGFGAVYKGYFADLD----LQVAVKKISRGSRQGKK 400
F+ +L ATNNF ++ +G G G VYKG F LD VAVK++ S G K
Sbjct: 216 CHHFSLADLKKATNNFDENNVIGDRGNGKVYKG-FLQLDHVATTVVAVKQMDNSSFWGLK 392
Query: 401 EYVTEVKVISQLRHRNLVKLLGWCHDKGEFLLVYEFMPNGSLDSHLFGK---RTPLSWGV 457
E+ E++++ QLRH NL+ L+G+C K E ++V+E+M NGSL L R PLSW
Sbjct: 393 EFKNEIELLCQLRHPNLITLIGFCLHKDEKIIVFEYMSNGSLADRLLNSRDARKPLSWKK 572
Query: 458 RHKIALGLASGLLYLHEEWERCVVHRDIKSSNVMLDSS 495
R +I +G A GL YLH +R + HR IK +N++LD +
Sbjct: 573 RIEICIGAARGLHYLHSGAKRNIFHRGIKPANILLDEN 686
>AI967315
Length = 1308
Score = 117 bits (293), Expect = 4e-27
Identities = 65/148 (43%), Positives = 95/148 (63%), Gaps = 3/148 (2%)
Frame = +1
Query: 348 FTYKELDLATNNFSKDRKLGQGGFGAVYKGYFADLDLQVAVKKISRGSRQGKKE--YVTE 405
F+Y+EL ATN FS + +G+GG+ VYKG D ++AVK+++R R +KE ++TE
Sbjct: 112 FSYEELFHATNGFSSENMVGKGGYAEVYKGRLESGD-EIAVKRLTRTCRDERKEKEFLTE 288
Query: 406 VKVISQLRHRNLVKLLGWCHDKGEFLLVYEFMPNGSLDSHLFG-KRTPLSWGVRHKIALG 464
+ I + H N++ LLG C D G +L V+E GS+ S + K PL W R+KI LG
Sbjct: 289 IGTIGHVCHSNVMPLLGCCIDNGLYL-VFELSTVGSVASLIHDEKMAPLDWKTRYKIVLG 465
Query: 465 LASGLLYLHEEWERCVVHRDIKSSNVML 492
A GL YLH+ +R ++HRDIK+SN++L
Sbjct: 466 TARGLHYLHKGCQRRIIHRDIKASNILL 549
>TC9462 similar to UP|O49840 (O49840) Protein kinase (AT2G02800/T20F6.6) ,
partial (37%)
Length = 576
Score = 117 bits (292), Expect = 6e-27
Identities = 69/154 (44%), Positives = 90/154 (57%), Gaps = 12/154 (7%)
Frame = +1
Query: 322 SNKQKEEAMHLTSMNDDLERGAGP--RRFTYKELDLATNNFSKDRKLGQGGFGAVYKGYF 379
S +K A L + + E + P + FT+ EL AT NF D LG+GGFG VYKG+
Sbjct: 112 SYSEKSNASSLPTPRSEGEILSSPNLKAFTFNELKNATRNFRPDSLLGEGGFGYVYKGWI 291
Query: 380 ADLD---------LQVAVKKISRGSRQGKKEYVTEVKVISQLRHRNLVKLLGWCHDKGEF 430
+ + VAVK++ QG KE++TEV + QL H NLVKL+G+C D
Sbjct: 292 DEHSFTAAKPGSGMVVAVKRLKPEGFQGHKEWLTEVNYLGQLYHPNLVKLIGYCLDGENR 471
Query: 431 LLVYEFMPNGSLDSHLFGK-RTPLSWGVRHKIAL 463
LLVYEFMP GSLD+HLF + PLSW R K+A+
Sbjct: 472 LLVYEFMPKGSLDNHLFTRGPQPLSWSXRXKLAI 573
>AV423294
Length = 459
Score = 117 bits (292), Expect = 6e-27
Identities = 61/129 (47%), Positives = 87/129 (67%), Gaps = 4/129 (3%)
Frame = +3
Query: 368 QGGFGAVYKGYFADLDLQVAVKKISRGSRQGKKEYVTEVKVISQLRHRNLVKLLGWCHDK 427
+GGFG VY+G+ ++ VA+K+++ QG +E+V EV +S H NLVKL+G+C +
Sbjct: 3 EGGFGKVYRGFIERINQVVAIKQLNPHGLQGIREFVVEVLTLSLADHPNLVKLIGFCAEG 182
Query: 428 GEFLLVYEFMPNGSLDSHLF----GKRTPLSWGVRHKIALGLASGLLYLHEEWERCVVHR 483
+ LLVYE+MP GSL+SHL GK+ PL W R KIA G A GL YLH + + V++R
Sbjct: 183 EQRLLVYEYMPLGSLESHLHDILPGKK-PLDWNTRMKIAAGAARGLEYLHNKMKPPVIYR 359
Query: 484 DIKSSNVML 492
DIK SN+++
Sbjct: 360 DIKCSNLLI 386
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.319 0.134 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,443,556
Number of Sequences: 28460
Number of extensions: 115613
Number of successful extensions: 935
Number of sequences better than 10.0: 210
Number of HSP's better than 10.0 without gapping: 796
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 798
length of query: 495
length of database: 4,897,600
effective HSP length: 94
effective length of query: 401
effective length of database: 2,222,360
effective search space: 891166360
effective search space used: 891166360
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 57 (26.6 bits)
Medicago: description of AC146709.1


