

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146708.4 + phase: 0
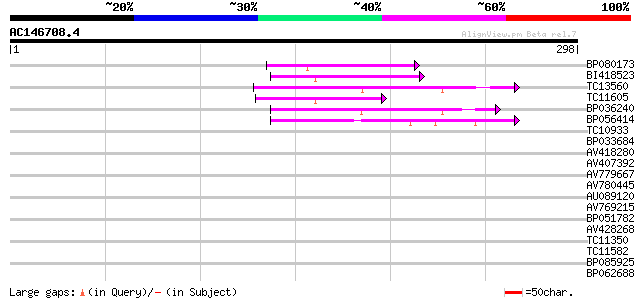
(298 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BP080173 52 2e-07
BI418523 47 5e-06
TC13560 46 9e-06
TC11605 41 2e-04
BP036240 40 4e-04
BP056414 40 5e-04
TC10933 similar to UP|Q9LNS0 (Q9LNS0) F1L3.4, partial (6%) 39 0.001
BP033684 37 0.005
AV418280 35 0.015
AV407392 34 0.034
AV779667 32 0.13
AV780445 30 0.49
AU089120 30 0.64
AV769215 29 0.84
BP051782 28 2.4
AV428268 27 3.2
TC11350 similar to UP|Q9LJP7 (Q9LJP7) Genomic DNA, chromosome 3,... 27 3.2
TC11582 similar to GB|AAP37821.1|30725598|BT008462 At3g18850 {Ar... 27 3.2
BP085925 27 4.2
BP062688 27 4.2
>BP080173
Length = 382
Score = 51.6 bits (122), Expect = 2e-07
Identities = 30/82 (36%), Positives = 41/82 (49%), Gaps = 2/82 (2%)
Frame = -2
Query: 136 VCWKPPSAPWVKVNTDGSVL--NNSGACGGLFRDHLGTFLGAFVGNLGRCSVFDTEVSGF 193
V W P W+K+N DGS +N ACGG+ DH GTF+ LG + E+ G
Sbjct: 351 VRWPVPMEGWIKINIDGSFAPSSNCAACGGVMWDHEGTFITGASVRLGCYFINHAELWGI 172
Query: 194 ILVMEHAALHGWYNIWLESDAS 215
+ A G+ I +ESD+S
Sbjct: 171 FHGAKIAQARGFKKILVESDSS 106
>BI418523
Length = 496
Score = 46.6 bits (109), Expect = 5e-06
Identities = 28/83 (33%), Positives = 41/83 (48%), Gaps = 2/83 (2%)
Frame = +1
Query: 138 WKPPSAPWVKVNTDGSVLNNSG--ACGGLFRDHLGTFLGAFVGNLGRCSVFDTEVSGFIL 195
W PPSA ++K N DG+ N G GG+FRD LG F N G ++ +V +
Sbjct: 187 WSPPSAGFIKCNVDGASQGNPGPSGVGGVFRDANRKILGYFSLNSGNGWAYEAKVRSILN 366
Query: 196 VMEHAALHGWYNIWLESDASSAL 218
+ NI +ESD++ A+
Sbjct: 367 ALVFIQKFLLKNILIESDSTVAV 435
>TC13560
Length = 571
Score = 45.8 bits (107), Expect = 9e-06
Identities = 44/153 (28%), Positives = 63/153 (40%), Gaps = 13/153 (8%)
Frame = +2
Query: 129 SMREIVSVCWKPPSAPWVKVNTDGSVLN-NSGACGGLFRDHLGTFLGAFVGNLGRCS--- 184
S + S W P A WVK+N DG+V N +C G+ R+ G++L F N G S
Sbjct: 119 SQTQNTSSDWIKPDAGWVKINVDGAVSNCLQASCRGVLRNADGSWLKGFRWNFGTFSSAN 298
Query: 185 VFDTEVSGFILVMEHAALHGWYNIWLESDASSALMVFKNPSL---------VPILLRNRW 235
VF TE+ +E A + +ESD+ + + L + IL R
Sbjct: 299 VFLTELMAVKTAVEAAMSLDLARVIVESDSLEVVDFLHSTDLGSHRYSQIALDILHLQRE 478
Query: 236 HNARTLNVQVISSHIFREGNVCVDRLANLGHSV 268
H A + GN VD +A G S+
Sbjct: 479 HGALVFQYAL-------RGNSIVDYIAKFGLSL 556
>TC11605
Length = 546
Score = 41.2 bits (95), Expect = 2e-04
Identities = 23/71 (32%), Positives = 33/71 (46%), Gaps = 2/71 (2%)
Frame = +3
Query: 130 MREIVSVCWKPPSAPWVKVNTDGSVLNNSG--ACGGLFRDHLGTFLGAFVGNLGRCSVFD 187
MR + + W PP A +K N DGS + G GG+ R+ LG F LG +
Sbjct: 324 MRMVRNCVWCPPEANILKFNVDGSARGSPGISGAGGILRNAESAVLGRFSKPLGVLCAYQ 503
Query: 188 TEVSGFILVME 198
EV +L ++
Sbjct: 504 AEVKAILLALK 536
>BP036240
Length = 567
Score = 40.4 bits (93), Expect = 4e-04
Identities = 34/134 (25%), Positives = 57/134 (42%), Gaps = 13/134 (9%)
Frame = -3
Query: 138 WKPPSAPWVKVNTDGSVLNNS-GACGGLFRDHLGTFLGAFVGNLGRC---SVFDTEVSGF 193
W P W K + DG++ + +CGG+ RD ++ F N+G +VF E+S
Sbjct: 562 WTAPDEGWTKFDVDGALSRDLLASCGGVLRDSRRHWIKGFCRNMGDLTYHNVFLVELSAI 383
Query: 194 ILVMEHAALHGWYNIWLESDASSALMVFKNPSL---------VPILLRNRWHNARTLNVQ 244
+E A ++ +ESDA A+ + + ++ IL R H +
Sbjct: 382 QSAVEIALSMDLQHVIVESDALEAIHLLSSENVHAHPFGSLAASILRMQRDHGS------ 221
Query: 245 VISSHIFREGNVCV 258
++ H RE N V
Sbjct: 220 IVFQHTPREANSLV 179
>BP056414
Length = 606
Score = 40.0 bits (92), Expect = 5e-04
Identities = 36/142 (25%), Positives = 61/142 (42%), Gaps = 11/142 (7%)
Frame = -3
Query: 138 WKPPSAPWVKVNTDGSVLNN-SGACGGLFRDHLGTFLGAFVGNLGRCSVFDTEVSGFILV 196
W P W KVN DG++ + +CGG+ RD G++ F +LG V + F+ +
Sbjct: 562 WTRPPEGWFKVNVDGALSGDLQASCGGVVRDAAGSWSKGFARSLG---VLRWHXAFFVEL 392
Query: 197 MEHAALHGWYNIW------LESDASSALMVFK--NPSLVPILLRNRWHNARTLNV--QVI 246
M + W +ESD+ + + + N S W + ++V ++
Sbjct: 391 MAVQTAVDFIMSWDIPQVIIESDSQQVVELLQNFNASSPHQYGEIAWDIQQKISVHGSIV 212
Query: 247 SSHIFREGNVCVDRLANLGHSV 268
+ RE N D LA +G S+
Sbjct: 211 ITSTPREANFLADYLAKVGLSL 146
>TC10933 similar to UP|Q9LNS0 (Q9LNS0) F1L3.4, partial (6%)
Length = 596
Score = 38.5 bits (88), Expect = 0.001
Identities = 17/29 (58%), Positives = 18/29 (61%), Gaps = 2/29 (6%)
Frame = +2
Query: 138 WKPPSAPWVKVNTDGSVLNNSG--ACGGL 164
W PPS WVK+NTDGS S CGGL
Sbjct: 107 WTPPSVGWVKLNTDGSFTRGSSLMGCGGL 193
>BP033684
Length = 557
Score = 36.6 bits (83), Expect = 0.005
Identities = 20/55 (36%), Positives = 27/55 (48%), Gaps = 2/55 (3%)
Frame = -1
Query: 138 WKPPSAPWVKVNTDGSVLNNSG--ACGGLFRDHLGTFLGAFVGNLGRCSVFDTEV 190
W PPS +K N DG+ N G GG+ D+ LG F N+G ++ EV
Sbjct: 287 WTPPSRGILKFNVDGASQGNPGPSGVGGVLCDYRSMVLGFFSINMGHGWAYEAEV 123
>AV418280
Length = 398
Score = 35.0 bits (79), Expect = 0.015
Identities = 27/86 (31%), Positives = 41/86 (47%), Gaps = 6/86 (6%)
Frame = -2
Query: 2 CSFCLEQVETSDHLFLRCKFVVTLWS----WLCSQLHVGLDFSSIK--ALMSSLPRHCSS 55
C FC E S HLF C F + +W WL + V L S++ A S R+ +
Sbjct: 388 CPFCTTVEECSGHLFFTCVFSMGVWQALHRWL--GISVALPASTLANFAQFSITARNKNQ 215
Query: 56 QVRDLYVAAVVHMVHSIWWARNNVRF 81
++ +L A + V S+W RN++ F
Sbjct: 214 RLGEL--AIWIATVWSLWIQRNSIIF 143
>AV407392
Length = 407
Score = 33.9 bits (76), Expect = 0.034
Identities = 13/25 (52%), Positives = 15/25 (60%)
Frame = -1
Query: 136 VCWKPPSAPWVKVNTDGSVLNNSGA 160
VCW P + WV +N DGS L N A
Sbjct: 254 VCWSSPPSSWVALNVDGSSLANQSA 180
>AV779667
Length = 455
Score = 32.0 bits (71), Expect = 0.13
Identities = 17/46 (36%), Positives = 22/46 (46%)
Frame = -3
Query: 243 VQVISSHIFREGNVCVDRLANLGHSVVGEVWLSTLPSDFHQVFYED 288
V + SSH+ REGN C D LA S VW+ P + +D
Sbjct: 186 VGIGSSHMGREGNSCADYLARNASSYPDSVWI*EGPPGLSSLLRDD 49
>AV780445
Length = 504
Score = 30.0 bits (66), Expect = 0.49
Identities = 16/38 (42%), Positives = 20/38 (52%), Gaps = 2/38 (5%)
Frame = +1
Query: 138 WKPPSAPWVKVNTDGS--VLNNSGACGGLFRDHLGTFL 173
W+PP VK+N DGS + GGL RD G +L
Sbjct: 391 WQPPRQGTVKLNVDGSGREQDRCMGAGGLIRDDQGKWL 504
>AU089120
Length = 586
Score = 29.6 bits (65), Expect = 0.64
Identities = 13/27 (48%), Positives = 15/27 (55%)
Frame = +2
Query: 1 MCSFCLEQVETSDHLFLRCKFVVTLWS 27
+ SFCL E+ HLF RC LWS
Sbjct: 35 LSSFCLHSEESIPHLFFRCFQSWNLWS 115
>AV769215
Length = 539
Score = 29.3 bits (64), Expect = 0.84
Identities = 12/29 (41%), Positives = 17/29 (58%)
Frame = -1
Query: 249 HIFREGNVCVDRLANLGHSVVGEVWLSTL 277
H++RE N D LA LG ++ +W TL
Sbjct: 524 HVYREANSLADHLAKLGCNLAIPIWGETL 438
>BP051782
Length = 523
Score = 27.7 bits (60), Expect = 2.4
Identities = 11/26 (42%), Positives = 17/26 (65%)
Frame = -2
Query: 249 HIFREGNVCVDRLANLGHSVVGEVWL 274
HI+REGN D+LA G + + +W+
Sbjct: 408 HIYREGNGEADKLAKEGCNRISNLWV 331
>AV428268
Length = 429
Score = 27.3 bits (59), Expect = 3.2
Identities = 10/27 (37%), Positives = 14/27 (51%)
Frame = +3
Query: 1 MCSFCLEQVETSDHLFLRCKFVVTLWS 27
+C C E++DHL C V +WS
Sbjct: 189 VCPLCSLDEESTDHLLFSCPIV*RIWS 269
>TC11350 similar to UP|Q9LJP7 (Q9LJP7) Genomic DNA, chromosome 3, P1 clone:
MIG10, partial (18%)
Length = 1118
Score = 27.3 bits (59), Expect = 3.2
Identities = 14/38 (36%), Positives = 21/38 (54%), Gaps = 2/38 (5%)
Frame = -3
Query: 138 WKPPSAPWVKVNTDGSVLNNSGAC--GGLFRDHLGTFL 173
W+PP K+N+D S S +C G + RD+LG +
Sbjct: 795 WRPPPLGVYKLNSDASWKAPSPSCTVGVVIRDNLGLLI 682
>TC11582 similar to GB|AAP37821.1|30725598|BT008462 At3g18850 {Arabidopsis
thaliana;}, partial (10%)
Length = 522
Score = 27.3 bits (59), Expect = 3.2
Identities = 13/28 (46%), Positives = 15/28 (53%)
Frame = +3
Query: 1 MCSFCLEQVETSDHLFLRCKFVVTLWSW 28
+CS L Q ET + LR K T WSW
Sbjct: 420 LCSCTL*QQETRVNRLLRLKCQYTFWSW 503
>BP085925
Length = 334
Score = 26.9 bits (58), Expect = 4.2
Identities = 13/36 (36%), Positives = 16/36 (44%)
Frame = -2
Query: 230 LLRNRWHNARTLNVQVISSHIFREGNVCVDRLANLG 265
L+ WH V+ +H REGN C D L G
Sbjct: 273 LMSRSWH--------VVLNHTLREGNACADFLTKHG 190
>BP062688
Length = 536
Score = 26.9 bits (58), Expect = 4.2
Identities = 13/27 (48%), Positives = 18/27 (66%)
Frame = -1
Query: 238 ARTLNVQVISSHIFREGNVCVDRLANL 264
AR+ NV + +H+ R+GN D LANL
Sbjct: 236 ARSWNVTI--NHVTRDGNAVADALANL 162
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.326 0.137 0.449
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,727,106
Number of Sequences: 28460
Number of extensions: 106145
Number of successful extensions: 769
Number of sequences better than 10.0: 55
Number of HSP's better than 10.0 without gapping: 764
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 765
length of query: 298
length of database: 4,897,600
effective HSP length: 90
effective length of query: 208
effective length of database: 2,336,200
effective search space: 485929600
effective search space used: 485929600
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 55 (25.8 bits)
Medicago: description of AC146708.4


