

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146708.15 - phase: 2
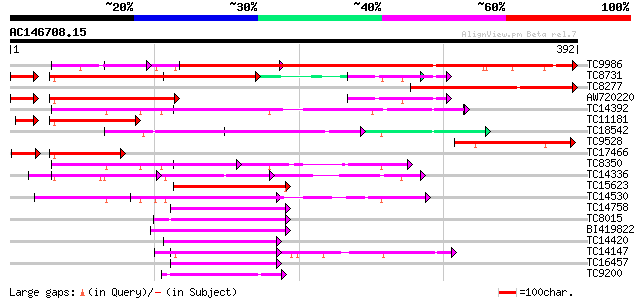
(392 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC9986 similar to UP|Q9M427 (Q9M427) Oligouridylate binding prot... 365 e-102
TC8731 homologue to UP|Q9LNS1 (Q9LNS1) F1L3.2, partial (34%) 292 5e-85
TC8277 similar to UP|Q9LJH8 (Q9LJH8) RNA binding protein nucleol... 214 3e-56
AW720220 182 4e-52
TC14392 similar to UP|Q9LEB4 (Q9LEB4) RNA binding protein 45, pa... 120 5e-28
TC11181 similar to UP|Q9M427 (Q9M427) Oligouridylate binding pro... 90 1e-20
TC18542 similar to UP|Q93YF1 (Q93YF1) Nucleic acid binding prote... 92 1e-19
TC9528 89 1e-18
TC17466 similar to UP|Q9M427 (Q9M427) Oligouridylate binding pro... 83 3e-18
TC8350 similar to UP|Q9LEB4 (Q9LEB4) RNA binding protein 45, par... 82 2e-16
TC14336 similar to UP|Q9M549 (Q9M549) Poly(A)-binding protein, p... 81 3e-16
TC15623 similar to UP|P93486 (P93486) Glycine-rich RNA-binding p... 70 5e-13
TC14530 similar to UP|P82277 (P82277) Plastid-specific 30S ribos... 65 1e-11
TC14758 similar to UP|O24601 (O24601) Glycine-rich RNA binding p... 65 2e-11
TC8015 similar to UP|ROC4_NICSY (P19683) 31 kDa ribonucleoprotei... 62 2e-10
BI419822 60 5e-10
TC14420 similar to UP|ROC3_NICSY (P19682) 28 kDa ribonucleoprote... 60 5e-10
TC14147 similar to UP|ROC1_NICSY (Q08935) 29 kDa ribonucleoprote... 58 2e-09
TC16457 similar to UP|Q41124 (Q41124) Chloroplast RNA binding pr... 57 4e-09
TC9200 similar to UP|Q8LLE6 (Q8LLE6) RNA-binding protein AKIP1, ... 56 9e-09
>TC9986 similar to UP|Q9M427 (Q9M427) Oligouridylate binding protein,
partial (46%)
Length = 1150
Score = 365 bits (936), Expect = e-102
Identities = 189/292 (64%), Positives = 228/292 (77%), Gaps = 17/292 (5%)
Frame = +3
Query: 118 DLSPEVTDATLFACFSVYQSCSDARVMWDQKTGRSRGFGFVSFRSQQDAQSAINDLTGKW 177
DLSPEVTDATLFACFSVY SCSDARVMWD KTGRS+G+GFVSFR QDAQS+IND+TGKW
Sbjct: 3 DLSPEVTDATLFACFSVYPSCSDARVMWDHKTGRSKGYGFVSFRDHQDAQSSINDMTGKW 182
Query: 178 LGSRQIRCNWATKGAGG-IEEKQNSDSKSVVELTNGSSEDGKEISSNDVPENNPQYTTVY 236
LG+RQIRCNWATKGAGG E++NSD+++ V LTNGSS+ G++ ++ + PENNP YTTVY
Sbjct: 183 LGNRQIRCNWATKGAGGSSSEEKNSDNQNAVVLTNGSSDGGQDNNNEEAPENNPSYTTVY 362
Query: 237 VGNLGSEATQLDLHRHFHALGAGVIEEVRVQRDKGFGFVRYSTHAEAALAIQMGNAQSYL 296
VGNL + TQ +LH FHALGAGVIEEVR+QRDKGFGFVRY+TH EAALAIQ+ N + +
Sbjct: 363 VGNLPHDVTQAELHCQFHALGAGVIEEVRIQRDKGFGFVRYNTHEEAALAIQVANGR-IV 539
Query: 297 CGKIIKCSWGSKPTPPGTASNPLPPPAAAP---LP------GLSATDILAYERQLAMSK- 346
GK +KCSWGS+PTPPGTASNPLPPPAA P LP G S ++LAY+RQLA+S+
Sbjct: 540 RGKTMKCSWGSRPTPPGTASNPLPPPAAQPYQILPTAGMNQGYSPAELLAYQRQLALSQA 719
Query: 347 --MGGVHAALMHPQAQHPLKQAAI----GASQAIYDGGFQNVAAAQQMMYYQ 392
G AL+ QH L A++ GASQA+YD G+ ++ QQ+MYY+
Sbjct: 720 AVSGLSGQALLQMTGQHGLAPASMGINTGASQAMYD-GYAGNSSRQQLMYYR 872
Score = 53.5 bits (127), Expect = 6e-08
Identities = 38/158 (24%), Positives = 67/158 (42%), Gaps = 33/158 (20%)
Frame = +3
Query: 66 YGFIHYFDRRSAALAILTLNGRHLFGQPIKVNWAY----ASGQREDTSGHYN-------- 113
YGF+ + D + A +I + G+ L + I+ NWA S E S + N
Sbjct: 114 YGFVSFRDHQDAQSSINDMTGKWLGNRQIRCNWATKGAGGSSSEEKNSDNQNAVVLTNGS 293
Query: 114 ---------------------IFVGDLSPEVTDATLFACFSVYQSCSDARVMWDQKTGRS 152
++VG+L +VT A L F A V+ + + R
Sbjct: 294 SDGGQDNNNEEAPENNPSYTTVYVGNLPHDVTQAELHCQFHAL----GAGVIEEVRIQRD 461
Query: 153 RGFGFVSFRSQQDAQSAINDLTGKWLGSRQIRCNWATK 190
+GFGFV + + ++A AI G+ + + ++C+W ++
Sbjct: 462 KGFGFVRYNTHEEAALAIQVANGRIVRGKTMKCSWGSR 575
Score = 42.4 bits (98), Expect = 1e-04
Identities = 26/71 (36%), Positives = 38/71 (52%), Gaps = 2/71 (2%)
Frame = +3
Query: 30 YVGNVHTQVTEPLLQEVF--AGTGPVEGCKLFRKEKSSYGFIHYFDRRSAALAILTLNGR 87
YVGN+ VT+ L F G G +E ++ R + +GF+ Y AALAI NGR
Sbjct: 360 YVGNLPHDVTQAELHCQFHALGAGVIEEVRIQRDK--GFGFVRYNTHEEAALAIQVANGR 533
Query: 88 HLFGQPIKVNW 98
+ G+ +K +W
Sbjct: 534 IVRGKTMKCSW 566
>TC8731 homologue to UP|Q9LNS1 (Q9LNS1) F1L3.2, partial (34%)
Length = 721
Score = 292 bits (747), Expect(2) = 5e-85
Identities = 143/148 (96%), Positives = 144/148 (96%), Gaps = 2/148 (1%)
Frame = +2
Query: 28 CR--YVGNVHTQVTEPLLQEVFAGTGPVEGCKLFRKEKSSYGFIHYFDRRSAALAILTLN 85
CR YVGN+HTQVTE LLQEVFAGTGPVEGCKLFRKEKSSYGFIHYFDRRSAALAILTLN
Sbjct: 278 CRSVYVGNIHTQVTELLLQEVFAGTGPVEGCKLFRKEKSSYGFIHYFDRRSAALAILTLN 457
Query: 86 GRHLFGQPIKVNWAYASGQREDTSGHYNIFVGDLSPEVTDATLFACFSVYQSCSDARVMW 145
GRHLFGQPIKVNWAYASGQREDTSGHYNIFVGDLSPEVTDATLFACFSVY SCSDARVMW
Sbjct: 458 GRHLFGQPIKVNWAYASGQREDTSGHYNIFVGDLSPEVTDATLFACFSVYPSCSDARVMW 637
Query: 146 DQKTGRSRGFGFVSFRSQQDAQSAINDL 173
DQKTGRSRGFGFVSFRSQQDAQSAINDL
Sbjct: 638 DQKTGRSRGFGFVSFRSQQDAQSAINDL 721
Score = 45.1 bits (105), Expect = 2e-05
Identities = 42/185 (22%), Positives = 74/185 (39%), Gaps = 4/185 (2%)
Frame = +2
Query: 107 DTSGHYNIFVGDLSPEVTDATLFACFSVYQSCSDARVMWDQKTGRSRGFGFVSFRSQQDA 166
D S +++VG++ +VT+ L F+ ++ +K+ +GF+ + ++ A
Sbjct: 266 DPSTCRSVYVGNIHTQVTELLLQEVFAGTGPVEGCKLFRKEKSS----YGFIHYFDRRSA 433
Query: 167 QSAINDLTGKWLGSRQIRCNWATKGAGGIEEKQNSDSKSVVELTNGSSEDGKEISSNDVP 226
AI L G+ L + I+ NWA +G ED
Sbjct: 434 ALAILTLNGRHLFGQPIKVNWA--------------------YASGQRED---------- 523
Query: 227 ENNPQYTTVYVGNLGSEATQLDLHRHFHAL----GAGVIEEVRVQRDKGFGFVRYSTHAE 282
+ ++VG+L E T L F A V+ + + R +GFGFV + + +
Sbjct: 524 --TSGHYNIFVGDLSPEVTDATLFACFSVYPSCSDARVMWDQKTGRSRGFGFVSFRSQQD 697
Query: 283 AALAI 287
A AI
Sbjct: 698 AQSAI 712
Score = 41.2 bits (95), Expect = 3e-04
Identities = 27/74 (36%), Positives = 42/74 (56%), Gaps = 2/74 (2%)
Frame = +2
Query: 234 TVYVGNLGSEATQLDLHRHFHALGAGVIEEVRVQRDK--GFGFVRYSTHAEAALAIQMGN 291
+VYVGN+ ++ T+L L F G G +E ++ R + +GF+ Y AALAI N
Sbjct: 284 SVYVGNIHTQVTELLLQEVF--AGTGPVEGCKLFRKEKSSYGFIHYFDRRSAALAILTLN 457
Query: 292 AQSYLCGKIIKCSW 305
+ +L G+ IK +W
Sbjct: 458 GR-HLFGQPIKVNW 496
Score = 39.7 bits (91), Expect(2) = 5e-85
Identities = 20/20 (100%), Positives = 20/20 (100%)
Frame = +3
Query: 1 LNQSQVEICLLVLIQVLAAV 20
LNQSQVEICLLVLIQVLAAV
Sbjct: 228 LNQSQVEICLLVLIQVLAAV 287
>TC8277 similar to UP|Q9LJH8 (Q9LJH8) RNA binding protein nucleolysin,
oligouridylate binding protein (AT3g14100/MAG2_5),
partial (26%)
Length = 789
Score = 214 bits (544), Expect = 3e-56
Identities = 106/115 (92%), Positives = 107/115 (92%)
Frame = +1
Query: 278 STHAEAALAIQMGNAQSYLCGKIIKCSWGSKPTPPGTASNPLPPPAAAPLPGLSATDILA 337
STHAEAALAIQMGN QS LCGK IKCSWGSKPTPPGTASNPLPPPA A LPG SATD+LA
Sbjct: 1 STHAEAALAIQMGNTQSILCGKQIKCSWGSKPTPPGTASNPLPPPAPATLPGFSATDLLA 180
Query: 338 YERQLAMSKMGGVHAALMHPQAQHPLKQAAIGASQAIYDGGFQNVAAAQQMMYYQ 392
YERQLAMSKMGGVH ALMHPQ QHPLKQAAIGASQAIYDGGFQNVAAAQQMMYYQ
Sbjct: 181 YERQLAMSKMGGVH-ALMHPQGQHPLKQAAIGASQAIYDGGFQNVAAAQQMMYYQ 342
>AW720220
Length = 535
Score = 182 bits (461), Expect(2) = 4e-52
Identities = 88/92 (95%), Positives = 89/92 (96%), Gaps = 2/92 (2%)
Frame = +3
Query: 28 CR--YVGNVHTQVTEPLLQEVFAGTGPVEGCKLFRKEKSSYGFIHYFDRRSAALAILTLN 85
CR YVGN+HTQVTE LLQEVFAGTGPVEGCKLFRKEKSSYGFIHYFDRRSAALAILTLN
Sbjct: 243 CRSVYVGNIHTQVTELLLQEVFAGTGPVEGCKLFRKEKSSYGFIHYFDRRSAALAILTLN 422
Query: 86 GRHLFGQPIKVNWAYASGQREDTSGHYNIFVG 117
GRHLFGQPIKVNWAYASGQREDTSGHYNIFVG
Sbjct: 423 GRHLFGQPIKVNWAYASGQREDTSGHYNIFVG 518
Score = 41.2 bits (95), Expect = 3e-04
Identities = 27/74 (36%), Positives = 42/74 (56%), Gaps = 2/74 (2%)
Frame = +3
Query: 234 TVYVGNLGSEATQLDLHRHFHALGAGVIEEVRVQRDK--GFGFVRYSTHAEAALAIQMGN 291
+VYVGN+ ++ T+L L F G G +E ++ R + +GF+ Y AALAI N
Sbjct: 249 SVYVGNIHTQVTELLLQEVF--AGTGPVEGCKLFRKEKSSYGFIHYFDRRSAALAILTLN 422
Query: 292 AQSYLCGKIIKCSW 305
+ +L G+ IK +W
Sbjct: 423 GR-HLFGQPIKVNW 461
Score = 39.7 bits (91), Expect(2) = 4e-52
Identities = 20/20 (100%), Positives = 20/20 (100%)
Frame = +1
Query: 1 LNQSQVEICLLVLIQVLAAV 20
LNQSQVEICLLVLIQVLAAV
Sbjct: 193 LNQSQVEICLLVLIQVLAAV 252
Score = 35.4 bits (80), Expect = 0.016
Identities = 21/82 (25%), Positives = 41/82 (49%)
Frame = +3
Query: 107 DTSGHYNIFVGDLSPEVTDATLFACFSVYQSCSDARVMWDQKTGRSRGFGFVSFRSQQDA 166
D S +++VG++ +VT+ L F+ ++ +K+ +GF+ + ++ A
Sbjct: 231 DPSTCRSVYVGNIHTQVTELLLQEVFAGTGPVEGCKLFRKEKSS----YGFIHYFDRRSA 398
Query: 167 QSAINDLTGKWLGSRQIRCNWA 188
AI L G+ L + I+ NWA
Sbjct: 399 ALAILTLNGRHLFGQPIKVNWA 464
>TC14392 similar to UP|Q9LEB4 (Q9LEB4) RNA binding protein 45, partial (77%)
Length = 1576
Score = 120 bits (300), Expect = 5e-28
Identities = 88/299 (29%), Positives = 147/299 (48%), Gaps = 10/299 (3%)
Frame = +3
Query: 30 YVGNVHTQVTEPLLQEVFAGTGPVEGCKLFRKEKSS----YGFIHYFDRRSAALAILTLN 85
++G++ + E L + FA TG + K+ R + ++ YGF+ + R A + T N
Sbjct: 342 WIGDLQYWMDENYLYQCFAHTGELATVKVIRNKNTNQSEGYGFLEFTSRAGAERILQTYN 521
Query: 86 GRHL--FGQPIKVNWAYASG---QREDTSGHYNIFVGDLSPEVTDATLFACFSV-YQSCS 139
G + GQ ++NWA S +R D S Y IFVGDL+ +VTD L F Y S
Sbjct: 522 GTIMPNGGQNFRLNWATFSAGGERRHDDSPDYTIFVGDLAADVTDYHLTEVFRTRYNSVK 701
Query: 140 DARVMWDQKTGRSRGFGFVSFRSQQDAQSAINDLTGKWLGSRQIRCNWATKGAGGIEEKQ 199
A+V+ D+ T R++G+GFV F + + A+ ++ G +R +R A
Sbjct: 702 GAKVVIDRLTSRTKGYGFVRFADESEQVRAMTEMQGVVCSTRPMRIGPAA---------- 851
Query: 200 NSDSKSVVELTNGSSEDGKEISSNDVPENNPQYTTVYVGNLGSEATQLDLHRHFHALGAG 259
+K++ T + + S EN+P TT++VGNL T L + F G
Sbjct: 852 ---NKNLGTGTGTQPKASYQNSQGPQNENDPNNTTIFVGNLDPNVTDDHLRQVFGLYGD- 1019
Query: 260 VIEEVRVQRDKGFGFVRYSTHAEAALAIQMGNAQSYLCGKIIKCSWGSKPTPPGTASNP 318
+ V++ + K GFV+++ + A A+++ N + L G+ ++ SWG P+ T S+P
Sbjct: 1020-LVHVKIPQGKRCGFVQFADRSCAEEALRVLNG-TLLGGQNVRLSWGRSPSNKQTQSDP 1190
Score = 70.5 bits (171), Expect = 5e-13
Identities = 56/211 (26%), Positives = 95/211 (44%), Gaps = 7/211 (3%)
Frame = +3
Query: 114 IFVGDLSPEVTDATLFACFSVYQSCSDARVMWDQKTGRSRGFGFVSFRSQQDAQSAINDL 173
+++GDL + + L+ CF+ + +V+ ++ T +S G+GF+ F S+ A+ +
Sbjct: 339 LWIGDLQYWMDENYLYQCFAHTGELATVKVIRNKNTNQSEGYGFLEFTSRAGAERILQTY 518
Query: 174 TGKWL--GSRQIRCNWATKGAGGIEEKQNSDSKSVVELTNGSSEDGKEISSNDVPENNPQ 231
G + G + R NWAT AGG E+++ DS P
Sbjct: 519 NGTIMPNGGQNFRLNWATFSAGG--ERRHDDS--------------------------PD 614
Query: 232 YTTVYVGNLGSEATQLDL-----HRHFHALGAGVIEEVRVQRDKGFGFVRYSTHAEAALA 286
Y T++VG+L ++ T L R+ GA V+ + R KG+GFVR++ +E A
Sbjct: 615 Y-TIFVGDLAADVTDYHLTEVFRTRYNSVKGAKVVIDRLTSRTKGYGFVRFADESEQVRA 791
Query: 287 IQMGNAQSYLCGKIIKCSWGSKPTPPGTASN 317
M Q +C ++P G A+N
Sbjct: 792 --MTEMQGVVC--------STRPMRIGPAAN 854
>TC11181 similar to UP|Q9M427 (Q9M427) Oligouridylate binding protein,
partial (24%)
Length = 499
Score = 90.1 bits (222), Expect(2) = 1e-20
Identities = 43/65 (66%), Positives = 50/65 (76%), Gaps = 2/65 (3%)
Frame = +2
Query: 28 CR--YVGNVHTQVTEPLLQEVFAGTGPVEGCKLFRKEKSSYGFIHYFDRRSAALAILTLN 85
CR YVGN+H VT+ LL EVF GP+ GCKL RKEKSSYGF+ Y DR SAALAI+TL+
Sbjct: 305 CRSVYVGNIHVNVTDKLLAEVFQSAGPLAGCKLIRKEKSSYGFVDYHDRASAALAIMTLH 484
Query: 86 GRHLF 90
GR L+
Sbjct: 485 GRQLY 499
Score = 30.0 bits (66), Expect = 0.69
Identities = 19/72 (26%), Positives = 34/72 (46%)
Frame = +2
Query: 107 DTSGHYNIFVGDLSPEVTDATLFACFSVYQSCSDARVMWDQKTGRSRGFGFVSFRSQQDA 166
D S +++VG++ VTD L F + +++ +K+ +GFV + + A
Sbjct: 293 DASSCRSVYVGNIHVNVTDKLLAEVFQSAGPLAGCKLIRKEKS----SYGFVDYHDRASA 460
Query: 167 QSAINDLTGKWL 178
AI L G+ L
Sbjct: 461 ALAIMTLHGRQL 496
Score = 29.6 bits (65), Expect = 0.90
Identities = 19/54 (35%), Positives = 26/54 (47%)
Frame = +2
Query: 234 TVYVGNLGSEATQLDLHRHFHALGAGVIEEVRVQRDKGFGFVRYSTHAEAALAI 287
+VYVGN+ T L F + G ++ + +GFV Y A AALAI
Sbjct: 311 SVYVGNIHVNVTDKLLAEVFQSAGPLAGCKLIRKEKSSYGFVDYHDRASAALAI 472
Score = 25.8 bits (55), Expect(2) = 1e-20
Identities = 12/16 (75%), Positives = 14/16 (87%)
Frame = +3
Query: 5 QVEICLLVLIQVLAAV 20
QVEICLLVL+ +LA V
Sbjct: 267 QVEICLLVLMLLLAVV 314
>TC18542 similar to UP|Q93YF1 (Q93YF1) Nucleic acid binding protein, partial
(29%)
Length = 568
Score = 92.0 bits (227), Expect = 1e-19
Identities = 62/184 (33%), Positives = 96/184 (51%), Gaps = 3/184 (1%)
Frame = +2
Query: 66 YGFIHYFDRRSAALAILTLNGRHLFG--QPIKVNWAYASGQREDTSGHYNIFVGDLSPEV 123
YGF+ + +A + T NG + G QP ++ WA +SG D ++IFVGDL+P+V
Sbjct: 23 YGFVEFVSHAAAERVLHTYNGTLMPGTEQPFRLIWA-SSGGGGDAGPDHSIFVGDLAPDV 199
Query: 124 TDATLFACFSV-YQSCSDARVMWDQKTGRSRGFGFVSFRSQQDAQSAINDLTGKWLGSRQ 182
TD L F Y S A+V+ D TGRS+G+GFV F + A+ ++ G + SR
Sbjct: 200 TDFLLQETFRTHYPSVRGAKVVTDPVTGRSKGYGFVKFSDEAQRNRAMTEMNGVYCSSRP 379
Query: 183 IRCNWATKGAGGIEEKQNSDSKSVVELTNGSSEDGKEISSNDVPENNPQYTTVYVGNLGS 242
+R + AT ++Q + +K++ + ++ S PEN+ TTV VGNL
Sbjct: 380 MRISAATPKKTPSFQQQYAPTKAMYQYPAYTAPVAP--VSVVAPENDVNNTTVCVGNLDL 553
Query: 243 EATQ 246
T+
Sbjct: 554 NVTE 565
Score = 43.5 bits (101), Expect = 6e-05
Identities = 44/191 (23%), Positives = 72/191 (37%), Gaps = 7/191 (3%)
Frame = +2
Query: 149 TGRSRGFGFVSFRSQQDAQSAINDLTGKWL-GSRQ-IRCNWATKGAGGIEEKQNSDSKSV 206
+ R+ G+GFV F S A+ ++ G + G+ Q R WA+ G GG
Sbjct: 5 SARAEGYGFVEFVSHAAAERVLHTYNGTLMPGTEQPFRLIWASSGGGG------------ 148
Query: 207 VELTNGSSEDGKEISSNDVPENNPQYTTVYVGNLGSEATQLDLHRHFHA-----LGAGVI 261
+ P ++ ++VG+L + T L F GA V+
Sbjct: 149 --------------------DAGPDHS-IFVGDLAPDVTDFLLQETFRTHYPSVRGAKVV 265
Query: 262 EEVRVQRDKGFGFVRYSTHAEAALAIQMGNAQSYLCGKIIKCSWGSKPTPPGTASNPLPP 321
+ R KG+GFV++S A+ A+ N Y + ++ S + P P
Sbjct: 266 TDPVTGRSKGYGFVKFSDEAQRNRAMTEMNG-VYCSSRPMRISAATPKKTPSFQQQYAPT 442
Query: 322 PAAAPLPGLSA 332
A P +A
Sbjct: 443 KAMYQYPAYTA 475
Score = 35.0 bits (79), Expect = 0.022
Identities = 22/75 (29%), Positives = 37/75 (49%), Gaps = 5/75 (6%)
Frame = +2
Query: 30 YVGNVHTQVTEPLLQEVFAGTGP-VEGCKLFRK----EKSSYGFIHYFDRRSAALAILTL 84
+VG++ VT+ LLQE F P V G K+ YGF+ + D A+ +
Sbjct: 173 FVGDLAPDVTDFLLQETFRTHYPSVRGAKVVTDPVTGRSKGYGFVKFSDEAQRNRAMTEM 352
Query: 85 NGRHLFGQPIKVNWA 99
NG + +P++++ A
Sbjct: 353 NGVYCSSRPMRISAA 397
>TC9528
Length = 548
Score = 89.4 bits (220), Expect = 1e-18
Identities = 49/91 (53%), Positives = 60/91 (65%), Gaps = 7/91 (7%)
Frame = +3
Query: 308 KPTPPGTASNPLP--PPAAAPLP-GLSATDILAYERQLAMSKMGGVHAALMHPQA-QHPL 363
KPTPPG AS+PLP PP+ AP+P G S + AYERQ+A+S+M G HAAL+ Q Q+
Sbjct: 3 KPTPPGAASSPLPLPPPSFAPVPAGFSLASLAAYERQMALSRMNGAHAALLQQQGYQNAA 182
Query: 364 KQAAIG---ASQAIYDGGFQNVAAAQQMMYY 391
KQAA+G A YD GFQ V Q +MYY
Sbjct: 183 KQAAMGMGALGAAAYDTGFQGVGTTQHLMYY 275
>TC17466 similar to UP|Q9M427 (Q9M427) Oligouridylate binding protein,
partial (20%)
Length = 389
Score = 83.2 bits (204), Expect(2) = 3e-18
Identities = 39/55 (70%), Positives = 45/55 (80%), Gaps = 2/55 (3%)
Frame = +2
Query: 28 CR--YVGNVHTQVTEPLLQEVFAGTGPVEGCKLFRKEKSSYGFIHYFDRRSAALA 80
CR YVGN+H QVT+ LLQE+FAG G +EGCKL RKEKSSYGF+ Y+DR SAA A
Sbjct: 224 CRSVYVGNIHPQVTDSLLQELFAGAGALEGCKLIRKEKSSYGFVDYYDRSSAAFA 388
Score = 30.8 bits (68), Expect = 0.41
Identities = 24/71 (33%), Positives = 35/71 (48%), Gaps = 4/71 (5%)
Frame = +2
Query: 220 ISSNDVPENNPQYT--TVYVGNLGSEATQLDLHRHFHALGAGVIEEVRVQRDK--GFGFV 275
I S ++P T +VYVGN+ + T L F GAG +E ++ R + +GFV
Sbjct: 182 IMSGNLPPGFDSSTCRSVYVGNIHPQVTDSLLQELF--AGAGALEGCKLIRKEKSSYGFV 355
Query: 276 RYSTHAEAALA 286
Y + AA A
Sbjct: 356 DYYDRSSAAFA 388
Score = 29.3 bits (64), Expect = 1.2
Identities = 16/63 (25%), Positives = 34/63 (53%)
Frame = +2
Query: 107 DTSGHYNIFVGDLSPEVTDATLFACFSVYQSCSDARVMWDQKTGRSRGFGFVSFRSQQDA 166
D+S +++VG++ P+VTD+ L F+ + +++ +K+ +GFV + + A
Sbjct: 212 DSSTCRSVYVGNIHPQVTDSLLQELFAGAGALEGCKLIRKEKS----SYGFVDYYDRSSA 379
Query: 167 QSA 169
A
Sbjct: 380 AFA 388
Score = 25.0 bits (53), Expect(2) = 3e-18
Identities = 14/20 (70%), Positives = 15/20 (75%)
Frame = +3
Query: 2 NQSQVEICLLVLIQVLAAVS 21
N S V ICLL LI+V AAVS
Sbjct: 177 NLS*VGICLLALIRVHAAVS 236
>TC8350 similar to UP|Q9LEB4 (Q9LEB4) RNA binding protein 45, partial (38%)
Length = 735
Score = 82.0 bits (201), Expect = 2e-16
Identities = 52/140 (37%), Positives = 78/140 (55%), Gaps = 9/140 (6%)
Frame = +1
Query: 30 YVGNVHTQVTEPLLQEVFAGTGPVEGCKLFRKEKSS----YGFIHYFDRRSAALAILTLN 85
++G++ + E L FA TG V K+ R +++S YGFI + R +A + T N
Sbjct: 307 WIGDLQYWMDENYLYTCFAHTGEVTTVKVIRNKQTSQSEGYGFIEFNSRAAAERVLQTFN 486
Query: 86 GRHL--FGQPIKVNWAYASG--QREDTSGHYNIFVGDLSPEVTDATLFACFSV-YQSCSD 140
G + GQ ++NWA S +R D + Y IFVGDL+ +VTD L F Y S
Sbjct: 487 GAIMPNGGQNFRLNWASLSAGEKRHDDTPDYTIFVGDLAADVTDYLLQETFRARYNSVKG 666
Query: 141 ARVMWDQKTGRSRGFGFVSF 160
A+V+ D+ +GR++G+GFV F
Sbjct: 667 AKVVIDRLSGRTKGYGFVRF 726
Score = 65.1 bits (157), Expect = 2e-11
Identities = 46/172 (26%), Positives = 81/172 (46%), Gaps = 7/172 (4%)
Frame = +1
Query: 114 IFVGDLSPEVTDATLFACFSVYQSCSDARVMWDQKTGRSRGFGFVSFRSQQDAQSAINDL 173
+++GDL + + L+ CF+ + +V+ +++T +S G+GF+ F S+ A+ +
Sbjct: 304 LWIGDLQYWMDENYLYTCFAHTGEVTTVKVIRNKQTSQSEGYGFIEFNSRAAAERVLQTF 483
Query: 174 TGKWL--GSRQIRCNWATKGAGGIEEKQNSDSKSVVELTNGSSEDGKEISSNDVPENNPQ 231
G + G + R NWA+ AG EK++ D+ P
Sbjct: 484 NGAIMPNGGQNFRLNWASLSAG---EKRHDDT--------------------------PD 576
Query: 232 YTTVYVGNLGSEATQLDLHRHFHA-----LGAGVIEEVRVQRDKGFGFVRYS 278
Y T++VG+L ++ T L F A GA V+ + R KG+GFVR++
Sbjct: 577 Y-TIFVGDLAADVTDYLLQETFRARYNSVKGAKVVIDRLSGRTKGYGFVRFA 729
>TC14336 similar to UP|Q9M549 (Q9M549) Poly(A)-binding protein, partial
(85%)
Length = 1917
Score = 80.9 bits (198), Expect = 3e-16
Identities = 77/289 (26%), Positives = 124/289 (42%), Gaps = 15/289 (5%)
Frame = +2
Query: 14 IQVLAAVSSLSVRKCR----YVGNVHTQVTEPLLQEVFAGTGPVEGCKLFRK---EKSSY 66
I+++ + S+RK ++ N+ + L + F+ G + CK+ + Y
Sbjct: 41 IRIMYSHRDPSIRKSGAGNIFIKNLDKAIDHKALHDTFSTFGNILSCKVATDSSGQSKGY 220
Query: 67 GFIHYFDRRSAALAILTLNGRHLFGQPIKVNWAYASGQRE---DTSGHYNIFVGDLSPEV 123
GF+ + +A AI LNG L + + V +RE D + N+FV +LS
Sbjct: 221 GFVQFETEEAAQNAIEKLNGMLLNDKQVYVGPFLRKQERESSTDKAKFNNVFVKNLSEST 400
Query: 124 TDATLFACFSVYQSCSDARVMWDQKTGRSRGFGFVSFRSQQDAQSAINDLTGKWLGSRQI 183
TD L F + + + A VM D G+S+ FGFV+F + DA A+ L GK + ++
Sbjct: 401 TDDELKNVFGEFGTITSAVVMRD-GDGKSKCFGFVNFENADDAARAVESLNGKKVDDKEW 577
Query: 184 RCNWATKGAGGIEEKQNSDSKSVVELTNGSSEDGKEISSNDVPENNPQYTTVYVGNLGSE 243
A K + +E +N +S+ E + Q +YV NL
Sbjct: 578 YVGKAQKKSEREQELKNKFEQSMKEAA-----------------DKYQGANLYVKNLDDS 706
Query: 244 ATQLDLHRHFHALGAGVIEEVRVQRD-----KGFGFVRYSTHAEAALAI 287
L F + G I +V RD +G GFV +ST EA+ A+
Sbjct: 707 IADEKLKELFSSY--GTITSCKVMRDPNGISRGSGFVAFSTPEEASRAL 847
Score = 61.6 bits (148), Expect = 2e-10
Identities = 45/172 (26%), Positives = 80/172 (46%), Gaps = 18/172 (10%)
Frame = +2
Query: 30 YVGNVHTQVTEPLLQEVFAGTGPVEGCKLFRK---EKSSYGFIHYFDRRSAALAILTLNG 86
+V N+ T+ L+ VF G + + R + +GF+++ + AA A+ +LNG
Sbjct: 374 FVKNLSESTTDDELKNVFGEFGTITSAVVMRDGDGKSKCFGFVNFENADDAARAVESLNG 553
Query: 87 RHLFGQPIKVNWAYASGQRE---------------DTSGHYNIFVGDLSPEVTDATLFAC 131
+ + + V A +RE D N++V +L + D L
Sbjct: 554 KKVDDKEWYVGKAQKKSEREQELKNKFEQSMKEAADKYQGANLYVKNLDDSIADEKLKEL 733
Query: 132 FSVYQSCSDARVMWDQKTGRSRGFGFVSFRSQQDAQSAINDLTGKWLGSRQI 183
FS Y + + +VM D G SRG GFV+F + ++A A+ ++ GK + S+ +
Sbjct: 734 FSSYGTITSCKVMRDPN-GISRGSGFVAFSTPEEASRALLEMNGKMVVSKPL 886
Score = 40.4 bits (93), Expect = 5e-04
Identities = 21/79 (26%), Positives = 38/79 (47%), Gaps = 3/79 (3%)
Frame = +2
Query: 30 YVGNVHTQVTEPLLQEVFAGTGPVEGCKLFRKEKS---SYGFIHYFDRRSAALAILTLNG 86
YV N+ + + L+E+F+ G + CK+ R GF+ + A+ A+L +NG
Sbjct: 683 YVKNLDDSIADEKLKELFSSYGTITSCKVMRDPNGISRGSGFVAFSTPEEASRALLEMNG 862
Query: 87 RHLFGQPIKVNWAYASGQR 105
+ + +P+ V A R
Sbjct: 863 KMVVSKPLYVTLAQRKEDR 919
Score = 26.2 bits (56), Expect = 10.0
Identities = 17/60 (28%), Positives = 27/60 (44%), Gaps = 3/60 (5%)
Frame = +2
Query: 235 VYVGNLGSEATQLDLHRHFHALGAGVIEEVRVQ---RDKGFGFVRYSTHAEAALAIQMGN 291
+++ NL LH F G + +V + KG+GFV++ T A AI+ N
Sbjct: 98 IFIKNLDKAIDHKALHDTFSTFGNILSCKVATDSSGQSKGYGFVQFETEEAAQNAIEKLN 277
>TC15623 similar to UP|P93486 (P93486) Glycine-rich RNA-binding protein
PsGRBP, partial (88%)
Length = 683
Score = 70.5 bits (171), Expect = 5e-13
Identities = 38/87 (43%), Positives = 56/87 (63%), Gaps = 6/87 (6%)
Frame = +1
Query: 114 IFVGDLSPEVTDATLFACFSVYQSCSDARVMWDQKTGRSRGFGFVSFRSQQDAQSAINDL 173
+F+G LS V D +L FS + + ++ARV+ D+ TGRSRGFGFVSF S++ A SA++ +
Sbjct: 190 LFIGGLSYGVDDKSLEDAFSSFGTVAEARVIVDRDTGRSRGFGFVSFDSEESASSALSSM 369
Query: 174 TGKWLGSRQIRCNWAT------KGAGG 194
G+ L R IR ++A +G GG
Sbjct: 370 DGQDLNGRNIRVSYANDRPAGPRGGGG 450
Score = 30.4 bits (67), Expect = 0.53
Identities = 23/83 (27%), Positives = 38/83 (45%), Gaps = 5/83 (6%)
Frame = +1
Query: 235 VYVGNLGSEATQLDLHRHFHALGAGVIEEVRVQRD----KGFGFVRYSTHAEAALAIQMG 290
+++G L L F + G V V RD +GFGFV + + A+ A+
Sbjct: 190 LFIGGLSYGVDDKSLEDAFSSFGTVAEARVIVDRDTGRSRGFGFVSFDSEESASSALSSM 369
Query: 291 NAQSYLCGKIIKCSWGS-KPTPP 312
+ Q L G+ I+ S+ + +P P
Sbjct: 370 DGQD-LNGRNIRVSYANDRPAGP 435
>TC14530 similar to UP|P82277 (P82277) Plastid-specific 30S ribosomal
protein 2, chloroplast precursor (PSRP-2), partial (68%)
Length = 1104
Score = 65.5 bits (158), Expect = 1e-11
Identities = 56/221 (25%), Positives = 97/221 (43%), Gaps = 13/221 (5%)
Frame = +2
Query: 84 LNGRHL-----FGQPIKVNWAYASGQRE---DTSGHYNIFVGDLSPEVTDATLFACFSVY 135
L RHL +P+++ +A ++ + ++VG++ +T+ L +
Sbjct: 122 LQPRHLRLAPSLARPLRLRRPFAVAEQAAATQSEADRRLYVGNIPRTLTNDELANIVQEH 301
Query: 136 QSCSDARVMWDQKTGRSRGFGFVSFRSQQDAQSAINDLTGKWLGSRQIRCNWATKGAGGI 195
+ A VM+D+ +GRSR F FV+ ++ +DA +AI L G +G R+I+ N K
Sbjct: 302 AAVEKAEVMYDKYSGRSRRFAFVTVKTVEDANAAIEKLNGTQIGGREIKVNVTEK----- 466
Query: 196 EEKQNSDSKSVVELTNGSSEDGKEISSNDVPENNPQYTTVYVGNLGSEATQLDLHRHFHA 255
+ V+ +E+ + + S VYVGNL T D+ ++F +
Sbjct: 467 -------PLTTVDFPLVQAEESEYVDSP---------YKVYVGNLAKTVTS-DMLKNFFS 595
Query: 256 -----LGAGVIEEVRVQRDKGFGFVRYSTHAEAALAIQMGN 291
L A V + GFGFV +S+ + AI N
Sbjct: 596 EKGKVLSAKVSRVPGTSKSSGFGFVTFSSDEDVEAAISSFN 718
Score = 62.8 bits (151), Expect = 1e-10
Identities = 52/186 (27%), Positives = 79/186 (41%), Gaps = 15/186 (8%)
Frame = +2
Query: 18 AAVSSLSVRKCRYVGNVHTQVTEPLLQEVFAGTGPVEGCKLFRKEKSS----YGFIHYFD 73
AA + + YVGN+ +T L + VE ++ + S + F+
Sbjct: 203 AAATQSEADRRLYVGNIPRTLTNDELANIVQEHAAVEKAEVMYDKYSGRSRRFAFVTVKT 382
Query: 74 RRSAALAILTLNGRHLFGQPIKVNWA-----------YASGQREDTSGHYNIFVGDLSPE 122
A AI LNG + G+ IKVN + + E Y ++VG+L+
Sbjct: 383 VEDANAAIEKLNGTQIGGREIKVNVTEKPLTTVDFPLVQAEESEYVDSPYKVYVGNLAKT 562
Query: 123 VTDATLFACFSVYQSCSDARVMWDQKTGRSRGFGFVSFRSQQDAQSAINDLTGKWLGSRQ 182
VT L FS A+V T +S GFGFV+F S +D ++AI+ L ++
Sbjct: 563 VTSDMLKNFFSEKGKVLSAKVSRVPGTSKSSGFGFVTFSSDEDVEAAISSFNNAPLEGQK 742
Query: 183 IRCNWA 188
IR N A
Sbjct: 743 IRVNKA 760
>TC14758 similar to UP|O24601 (O24601) Glycine-rich RNA binding protein 2,
partial (93%)
Length = 725
Score = 65.1 bits (157), Expect = 2e-11
Identities = 36/83 (43%), Positives = 47/83 (56%)
Frame = +1
Query: 112 YNIFVGDLSPEVTDATLFACFSVYQSCSDARVMWDQKTGRSRGFGFVSFRSQQDAQSAIN 171
Y FVG L+ TL FS Y D++V+ D++TGRSRGFGFV+F S++ +SAI
Sbjct: 82 YRCFVGGLAWTTDSHTLEQAFSSYGEIIDSKVVNDRETGRSRGFGFVTFTSEEAMRSAIE 261
Query: 172 DLTGKWLGSRQIRCNWATKGAGG 194
+ G L R I N A GG
Sbjct: 262 GMNGNELDGRNITVNEAQARGGG 330
Score = 32.0 bits (71), Expect = 0.18
Identities = 20/78 (25%), Positives = 35/78 (44%), Gaps = 4/78 (5%)
Frame = +1
Query: 30 YVGNVHTQVTEPLLQEVFAGTGPVEGCKLFRKEKSS----YGFIHYFDRRSAALAILTLN 85
+VG + L++ F+ G + K+ ++ +GF+ + + AI +N
Sbjct: 91 FVGGLAWTTDSHTLEQAFSSYGEIIDSKVVNDRETGRSRGFGFVTFTSEEAMRSAIEGMN 270
Query: 86 GRHLFGQPIKVNWAYASG 103
G L G+ I VN A A G
Sbjct: 271 GNELDGRNITVNEAQARG 324
>TC8015 similar to UP|ROC4_NICSY (P19683) 31 kDa ribonucleoprotein,
chloroplast precursor, partial (28%)
Length = 717
Score = 62.0 bits (149), Expect = 2e-10
Identities = 37/95 (38%), Positives = 54/95 (55%)
Frame = +3
Query: 100 YASGQREDTSGHYNIFVGDLSPEVTDATLFACFSVYQSCSDARVMWDQKTGRSRGFGFVS 159
+ SG SG +++VG+L V A L F + + +AR++ D++TGRSRGFGFV+
Sbjct: 45 FGSGSGSRDSG-LSVYVGNLPWSVDTARLEEIFREHGNVENARIVMDRETGRSRGFGFVT 221
Query: 160 FRSQQDAQSAINDLTGKWLGSRQIRCNWATKGAGG 194
S+ D AI L G+ L R IR + A +GG
Sbjct: 222 MSSEADINGAIAALDGQSLDGRTIRVSVAEGRSGG 326
Score = 34.7 bits (78), Expect = 0.028
Identities = 26/83 (31%), Positives = 40/83 (47%), Gaps = 4/83 (4%)
Frame = +3
Query: 21 SSLSVRKCRYVGNVHTQVTEPLLQEVFAGTGPVEGCKLFRKEKS----SYGFIHYFDRRS 76
S LSV YVGN+ V L+E+F G VE ++ ++ +GF+
Sbjct: 72 SGLSV----YVGNLPWSVDTARLEEIFREHGNVENARIVMDRETGRSRGFGFVTMSSEAD 239
Query: 77 AALAILTLNGRHLFGQPIKVNWA 99
AI L+G+ L G+ I+V+ A
Sbjct: 240 INGAIAALDGQSLDGRTIRVSVA 308
Score = 32.7 bits (73), Expect = 0.11
Identities = 26/75 (34%), Positives = 36/75 (47%), Gaps = 4/75 (5%)
Frame = +3
Query: 234 TVYVGNLGSEATQLDLHRHFHALG----AGVIEEVRVQRDKGFGFVRYSTHAEAALAIQM 289
+VYVGNL L F G A ++ + R +GFGFV S+ A+ AI
Sbjct: 81 SVYVGNLPWSVDTARLEEIFREHGNVENARIVMDRETGRSRGFGFVTMSSEADINGAIAA 260
Query: 290 GNAQSYLCGKIIKCS 304
+ QS L G+ I+ S
Sbjct: 261 LDGQS-LDGRTIRVS 302
>BI419822
Length = 380
Score = 60.5 bits (145), Expect = 5e-10
Identities = 34/98 (34%), Positives = 53/98 (53%), Gaps = 1/98 (1%)
Frame = +2
Query: 98 WAYASGQREDTSGHYNIFVGDLSPEVTDATLFACFSVYQSCSDARVMWDQKTGRSRGFGF 157
W ++ + FVG L+ + L FS Y +++V+ D++TGRSRGFGF
Sbjct: 35 WFFSFSSMASAEIEFRCFVGGLAWATDNEALEKAFSPYGEIVESKVINDRETGRSRGFGF 214
Query: 158 VSFRSQQDAQSAINDLTGKWLGSRQIRCNWA-TKGAGG 194
V+F S+Q + AI + G+ L R I N A ++G+ G
Sbjct: 215 VTFASEQAMKDAIEAMNGQNLDGRNITVNEAQSRGSSG 328
Score = 32.0 bits (71), Expect = 0.18
Identities = 20/90 (22%), Positives = 44/90 (48%), Gaps = 4/90 (4%)
Frame = +2
Query: 18 AAVSSLSVRKCRYVGNVHTQVTEPLLQEVFAGTGPVEGCKLFRKEKSS----YGFIHYFD 73
++++S + +VG + L++ F+ G + K+ ++ +GF+ +
Sbjct: 50 SSMASAEIEFRCFVGGLAWATDNEALEKAFSPYGEIVESKVINDRETGRSRGFGFVTFAS 229
Query: 74 RRSAALAILTLNGRHLFGQPIKVNWAYASG 103
++ AI +NG++L G+ I VN A + G
Sbjct: 230 EQAMKDAIEAMNGQNLDGRNITVNEAQSRG 319
Score = 26.9 bits (58), Expect = 5.9
Identities = 18/63 (28%), Positives = 28/63 (43%), Gaps = 4/63 (6%)
Frame = +2
Query: 236 YVGNLGSEATQLDLHRHFHALG----AGVIEEVRVQRDKGFGFVRYSTHAEAALAIQMGN 291
+VG L L + F G + VI + R +GFGFV +++ AI+ N
Sbjct: 86 FVGGLAWATDNEALEKAFSPYGEIVESKVINDRETGRSRGFGFVTFASEQAMKDAIEAMN 265
Query: 292 AQS 294
Q+
Sbjct: 266 GQN 274
>TC14420 similar to UP|ROC3_NICSY (P19682) 28 kDa ribonucleoprotein,
chloroplast precursor (28RNP), partial (34%)
Length = 637
Score = 60.5 bits (145), Expect = 5e-10
Identities = 32/82 (39%), Positives = 48/82 (58%)
Frame = +1
Query: 107 DTSGHYNIFVGDLSPEVTDATLFACFSVYQSCSDARVMWDQKTGRSRGFGFVSFRSQQDA 166
D + ++VG+L + ++ L FS + ARV++D++TGRSRGFGFV+ + +
Sbjct: 25 DAQPAFRVYVGNLPWDFDNSQLEQVFSEHGKVVSARVVYDRETGRSRGFGFVTMSDEAEL 204
Query: 167 QSAINDLTGKWLGSRQIRCNWA 188
AI L G+ LG R IR N A
Sbjct: 205 NDAIAALDGQSLGGRTIRVNVA 270
Score = 33.1 bits (74), Expect = 0.082
Identities = 26/72 (36%), Positives = 35/72 (48%), Gaps = 4/72 (5%)
Frame = +1
Query: 235 VYVGNLGSEATQLDLHR----HFHALGAGVIEEVRVQRDKGFGFVRYSTHAEAALAIQMG 290
VYVGNL + L + H + A V+ + R +GFGFV S AE AI
Sbjct: 46 VYVGNLPWDFDNSQLEQVFSEHGKVVSARVVYDRETGRSRGFGFVTMSDEAELNDAIAAL 225
Query: 291 NAQSYLCGKIIK 302
+ QS L G+ I+
Sbjct: 226 DGQS-LGGRTIR 258
>TC14147 similar to UP|ROC1_NICSY (Q08935) 29 kDa ribonucleoprotein A,
chloroplast precursor (CP29A), partial (63%)
Length = 1602
Score = 58.2 bits (139), Expect = 2e-09
Identities = 35/91 (38%), Positives = 52/91 (56%), Gaps = 3/91 (3%)
Frame = +3
Query: 101 ASGQREDTSGHYN---IFVGDLSPEVTDATLFACFSVYQSCSDARVMWDQKTGRSRGFGF 157
+ G R S Y+ + VG+L+ V +A L + F DA+V++D+++GRSRGFGF
Sbjct: 696 SDGYRGGGSSSYSENRVHVGNLAWGVDNAALESLFREQGRVVDAKVIYDRESGRSRGFGF 875
Query: 158 VSFRSQQDAQSAINDLTGKWLGSRQIRCNWA 188
V+F S + SAI L G L R I+ + A
Sbjct: 876 VTFSSPDEVNSAIRSLDGADLNGRAIKVSQA 968
Score = 58.2 bits (139), Expect = 2e-09
Identities = 65/218 (29%), Positives = 92/218 (41%), Gaps = 20/218 (9%)
Frame = +3
Query: 112 YNIFVGDLSPEVTDATLFACFSVYQSCSDARVMWDQKTGRSRGFGFVSFRSQQDAQSAIN 171
+ +FVG+L V A L F + V++D+ TG SRGF FV+ S +A++A
Sbjct: 345 HKVFVGNLPFSVDSAQLAELFQDAGNVEVVEVIYDKMTGNSRGFAFVTMSSAAEAEAAAQ 524
Query: 172 DLTGKWLGSRQIRCNWATKGAG---GIEE----KQNSDSKSVVELTNGSSE-------DG 217
L R +R N G E + NS ++ + G S+ DG
Sbjct: 525 QFNNYELEGRALRVNSGPPPKNENRGFNENPRFRNNSFNRGGSDSYRGGSDGYRGGGSDG 704
Query: 218 -KEISSNDVPENNPQYTTVYVGNLGSEATQLDLHRHFHALG----AGVIEEVRVQRDKGF 272
+ S+ EN V+VGNL L F G A VI + R +GF
Sbjct: 705 YRGGGSSSYSENR-----VHVGNLAWGVDNAALESLFREQGRVVDAKVIYDRESGRSRGF 869
Query: 273 GFVRYSTHAEAALAIQMGNAQSYLCGKIIKCSWG-SKP 309
GFV +S+ E AI+ + L G+ IK S SKP
Sbjct: 870 GFVTFSSPDEVNSAIRSLDGAD-LNGRAIKVSQADSKP 980
Score = 33.9 bits (76), Expect = 0.048
Identities = 24/81 (29%), Positives = 39/81 (47%), Gaps = 4/81 (4%)
Frame = +3
Query: 30 YVGNVHTQVTEPLLQEVFAGTGPVEGCKLFRKEKSS----YGFIHYFDRRSAALAILTLN 85
+VGN+ V L+ +F G V K+ +S +GF+ + AI +L+
Sbjct: 747 HVGNLAWGVDNAALESLFREQGRVVDAKVIYDRESGRSRGFGFVTFSSPDEVNSAIRSLD 926
Query: 86 GRHLFGQPIKVNWAYASGQRE 106
G L G+ IKV+ A + +RE
Sbjct: 927 GADLNGRAIKVSQADSKPKRE 989
>TC16457 similar to UP|Q41124 (Q41124) Chloroplast RNA binding protein
precursor, partial (30%)
Length = 655
Score = 57.4 bits (137), Expect = 4e-09
Identities = 30/77 (38%), Positives = 46/77 (58%)
Frame = +2
Query: 112 YNIFVGDLSPEVTDATLFACFSVYQSCSDARVMWDQKTGRSRGFGFVSFRSQQDAQSAIN 171
+ +FVG+LS VT +L F Y + ARV++D +TGRSRG+GFV + + + ++A+
Sbjct: 14 HKLFVGNLSWTVTSESLIQVFPEYGTVVGARVLYDGETGRSRGYGFVCYSKRSELETALI 193
Query: 172 DLTGKWLGSRQIRCNWA 188
L L R IR + A
Sbjct: 194 SLNNVELEGRAIRVSLA 244
>TC9200 similar to UP|Q8LLE6 (Q8LLE6) RNA-binding protein AKIP1, partial
(31%)
Length = 727
Score = 56.2 bits (134), Expect = 9e-09
Identities = 31/86 (36%), Positives = 50/86 (58%)
Frame = +1
Query: 106 EDTSGHYNIFVGDLSPEVTDATLFACFSVYQSCSDARVMWDQKTGRSRGFGFVSFRSQQD 165
ED S H IFV L + T TL F Y + D + + D+ TG+S+G+GF+ F+S++
Sbjct: 451 EDVS-HRKIFVHGLGWDTTATTLVYAFQQYGAIEDCKAVTDKVTGKSKGYGFILFKSRRG 627
Query: 166 AQSAINDLTGKWLGSRQIRCNWATKG 191
A++A+ + K +G+R C A+ G
Sbjct: 628 ARNALKE-PQKKIGNRMTACQLASIG 702
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.317 0.132 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,683,217
Number of Sequences: 28460
Number of extensions: 94460
Number of successful extensions: 1562
Number of sequences better than 10.0: 164
Number of HSP's better than 10.0 without gapping: 1236
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1497
length of query: 392
length of database: 4,897,600
effective HSP length: 92
effective length of query: 300
effective length of database: 2,279,280
effective search space: 683784000
effective search space used: 683784000
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 56 (26.2 bits)
Medicago: description of AC146708.15


