

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146683.4 + phase: 0 /pseudo
(424 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
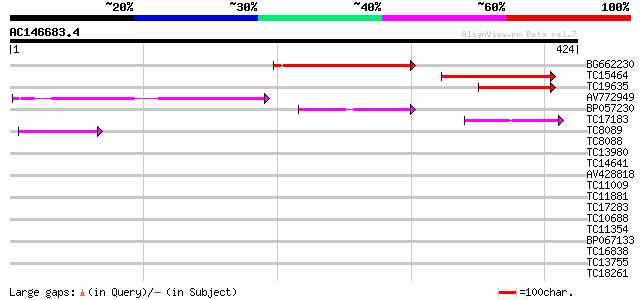
Score E
Sequences producing significant alignments: (bits) Value
BG662230 193 4e-50
TC15464 160 3e-40
TC19635 114 4e-26
AV772949 82 2e-16
BP057230 57 6e-09
TC17183 weakly similar to UP|Q95RC5 (Q95RC5) LD44506p (CG5745-PA... 44 7e-05
TC8089 similar to UP|FLA1_ARATH (Q9FM65) Fasciclin-like arabinog... 40 6e-04
TC8088 similar to UP|Q9C5Q6 (Q9C5Q6) Fasciclin-like arabinogalac... 39 0.002
TC13980 similar to UP|Q9SGT0 (Q9SGT0) T6H22.18 protein (F14J16.3... 38 0.003
TC14641 similar to UP|NTF2_ARATH (Q9C7F5) Nuclear transport fact... 38 0.003
AV428818 38 0.004
TC11009 similar to GB|AAP21377.1|30102918|BT006569 At1g47970 {Ar... 37 0.005
TC11881 similar to PIR|T48303|T48303 meiosis specific-like prote... 37 0.006
TC17283 similar to UP|O65760 (O65760) Extensin (Fragment), parti... 37 0.008
TC10688 similar to UP|AAR10812 (AAR10812) Superoxide dismutase, ... 37 0.008
TC11354 similar to UP|CAE54480 (CAE54480) Alpha glucosidase II ... 37 0.008
BP067133 36 0.011
TC16838 similar to UP|BAD07764 (BAD07764) GTPase activating prot... 36 0.011
TC13755 similar to UP|Q8HCN7 (Q8HCN7) Ribosomal protein L2, part... 36 0.011
TC18261 similar to UP|WSC2_YEAST (P53832) Cell wall integrity an... 36 0.011
>BG662230
Length = 340
Score = 193 bits (491), Expect = 4e-50
Identities = 92/106 (86%), Positives = 99/106 (92%)
Frame = +3
Query: 198 DDVKCETRGMLSRSQITHGEHPLSLGKTSIWNQFFQDTDIIEQIDRDVKRTHPDMHFFCG 257
D+ KCE R +LSRS+ITHGEHPLSLGKTS+WNQFFQDT+II+QIDRDV RTHPDMHFF G
Sbjct: 15 DEAKCE-RALLSRSEITHGEHPLSLGKTSVWNQFFQDTEIIDQIDRDVMRTHPDMHFFSG 191
Query: 258 DSQLAKSNQEALKNILIIFAKLNPGIRYVQGMNEVLAPLFYVFKND 303
DSQ AK NQEALKNILIIFAKLNPG+RYVQGMNEVLAPLFYVFKND
Sbjct: 192 DSQFAKLNQEALKNILIIFAKLNPGVRYVQGMNEVLAPLFYVFKND 329
>TC15464
Length = 672
Score = 160 bits (406), Expect = 3e-40
Identities = 74/85 (87%), Positives = 82/85 (96%)
Frame = +2
Query: 324 LSGFRDNFCQQLDNSIVGIRSTITRLSQLLKEHDEELWRHLEVTTKVNPQFYAFRWITLL 383
LSGFRDNF QQLDNS VGIR+TITRLSQLLKEHDEELWRHLE+T+KVNPQFYAFRWITLL
Sbjct: 2 LSGFRDNFVQQLDNSPVGIRATITRLSQLLKEHDEELWRHLEITSKVNPQFYAFRWITLL 181
Query: 384 LTQEFDFADSLRIWDTLVSDPDGPQ 408
LTQEF+FADS+ IWDT++SDP+GPQ
Sbjct: 182 LTQEFNFADSIHIWDTILSDPEGPQ 256
>TC19635
Length = 566
Score = 114 bits (284), Expect = 4e-26
Identities = 51/58 (87%), Positives = 55/58 (93%)
Frame = +3
Query: 351 QLLKEHDEELWRHLEVTTKVNPQFYAFRWITLLLTQEFDFADSLRIWDTLVSDPDGPQ 408
QLLKEHDEELWRHLEVTTKVNPQFY+FRWITLLLTQEF+F D L IWDTL+SDP+GPQ
Sbjct: 3 QLLKEHDEELWRHLEVTTKVNPQFYSFRWITLLLTQEFNFHDILHIWDTLLSDPEGPQ 176
>AV772949
Length = 521
Score = 82.0 bits (201), Expect = 2e-16
Identities = 68/193 (35%), Positives = 93/193 (47%), Gaps = 1/193 (0%)
Frame = +1
Query: 3 KKRVPDWLNSPMWSSPTEQNRFSATSYPSEPPSPPPVTVVEDPPSIHNAITTSPPSSSSS 62
+K+VP +NS +WSSP + PPS + TTS PS
Sbjct: 25 EKKVP-MVNSSLWSSPI-----------TTPPSSGRGRRHPRRRHLTGTATTSFPSRHHP 168
Query: 63 SASTSSTDDMSISRLAQSQSQLLAEVPFTLKSKCL*LLFSIQLY*YC*LFVY**LSRKVI 122
+ST + + R + LL+ +L L + +SRKVI
Sbjct: 169 RSSTGTENPGLAIRRVTATHPLLSPPAMSLTR----------------LRWWAEMSRKVI 300
Query: 123 DMRELRKIASQGIPDSP-GLRSTIWKLLLGYLPPDRSLWSSELAKKRSQYKRFKQDILIN 181
D+RELRK G L LGYLPPDR L SSELAKKR+QYK++K+++L+N
Sbjct: 301 DIRELRKRRV*WCT*CVLGYAPACGSLWLGYLPPDRGLRSSELAKKRTQYKQYKEEVLMN 480
Query: 182 PSEITRRMFNSAS 194
PSE+T +M+NS S
Sbjct: 481 PSELTWKMYNSLS 519
>BP057230
Length = 541
Score = 57.0 bits (136), Expect = 6e-09
Identities = 29/87 (33%), Positives = 50/87 (57%)
Frame = -2
Query: 217 EHPLSLGKTSIWNQFFQDTDIIEQIDRDVKRTHPDMHFFCGDSQLAKSNQEALKNILIIF 276
++PLS S W++FF++ ++ +D+D+ R +P+ G Q L+ IL+++
Sbjct: 426 DNPLSQNPDSTWSRFFRNAELERMVDQDLSRLYPEH----GSYFQTPGCQGMLRRILLLW 259
Query: 277 AKLNPGIRYVQGMNEVLAPLFYVFKND 303
+P Y QGM+E+LAPL YV + D
Sbjct: 258 CLRHPECGYRQGMHELLAPLLYVLQVD 178
>TC17183 weakly similar to UP|Q95RC5 (Q95RC5) LD44506p (CG5745-PA), partial
(6%)
Length = 678
Score = 43.5 bits (101), Expect = 7e-05
Identities = 25/75 (33%), Positives = 40/75 (53%), Gaps = 1/75 (1%)
Frame = +1
Query: 341 GIRSTITRLSQLLKEHDEELWRHLEVTTKVNPQFYAFRWITLLLTQEFDFADSLRIWDTL 400
GI+ + +L +L++ D+ + H+E QF AFRW LL +E F R+WDT
Sbjct: 7 GIQRLVFKLKELVRRIDDPVSTHMENQGLEFLQF-AFRWFNCLLIREIPFNMVTRLWDTY 183
Query: 401 VSDPDG-PQVQIYSF 414
+++ D P +Y F
Sbjct: 184 LAEGDALPDFLVYIF 228
>TC8089 similar to UP|FLA1_ARATH (Q9FM65) Fasciclin-like arabinogalactan
protein 1 precursor, partial (43%)
Length = 890
Score = 40.4 bits (93), Expect = 6e-04
Identities = 24/63 (38%), Positives = 30/63 (47%)
Frame = +2
Query: 7 PDWLNSPMWSSPTEQNRFSATSYPSEPPSPPPVTVVEDPPSIHNAITTSPPSSSSSSAST 66
P +SP SS T S+T P+ PPSPP T PP +TSP S++ S S
Sbjct: 398 PSRTSSPSTSSSTTSAPRSSTRSPTAPPSPPQCTKPPAPPRAPPDSSTSPTSAAGKSDSA 577
Query: 67 SST 69
T
Sbjct: 578 PRT 586
Score = 30.4 bits (67), Expect = 0.58
Identities = 22/81 (27%), Positives = 32/81 (39%), Gaps = 9/81 (11%)
Frame = +2
Query: 11 NSPMWSSPTEQNRFSATSYPSEPPSPPPVTVVEDP---------PSIHNAITTSPPSSSS 61
+S W P + S PS P SPP T P P +A +T+PP ++S
Sbjct: 194 SSSPWLLPPQTPTTSPAFSPSTPSSPPSTTTSPSPTSPPRSTNGPQSPSAPSTTPPWTTS 373
Query: 62 SSASTSSTDDMSISRLAQSQS 82
S + S + S S +
Sbjct: 374 SQNTHQSPPSRTSSPSTSSST 436
Score = 30.4 bits (67), Expect = 0.58
Identities = 26/68 (38%), Positives = 34/68 (49%), Gaps = 10/68 (14%)
Frame = +2
Query: 12 SPMWSSPTEQNRFSATSYPSEPPSPPPVTVVEDPPSIHNAI---TTS-------PPSSSS 61
SP +S T + S T+ PS P SPP T PS + TTS PPS +S
Sbjct: 236 SPAFSPSTPSSPPSTTTSPS-PTSPPRSTNGPQSPSAPSTTPPWTTSSQNTHQSPPSRTS 412
Query: 62 SSASTSST 69
S +++SST
Sbjct: 413 SPSTSSST 436
Score = 27.3 bits (59), Expect = 4.9
Identities = 25/69 (36%), Positives = 32/69 (46%), Gaps = 6/69 (8%)
Frame = +2
Query: 7 PDWLNSPMWSSPTEQNRFSATSYPSEPPSPPPVTVV-----EDPPSIHNAITTSPPSSSS 61
P SP +SP + PS P + PP T + PPS T+SP +SSS
Sbjct: 272 PSTTTSPSPTSPPRST--NGPQSPSAPSTTPPWTTSSQNTHQSPPSR----TSSPSTSSS 433
Query: 62 -SSASTSST 69
+SA SST
Sbjct: 434 TTSAPRSST 460
>TC8088 similar to UP|Q9C5Q6 (Q9C5Q6) Fasciclin-like
arabinogalactan-protein 2, partial (69%)
Length = 1527
Score = 38.5 bits (88), Expect = 0.002
Identities = 25/63 (39%), Positives = 31/63 (48%)
Frame = +2
Query: 11 NSPMWSSPTEQNRFSATSYPSEPPSPPPVTVVEDPPSIHNAITTSPPSSSSSSASTSSTD 70
+SP SS T S+T P+EPPS PP + PP A +TSP S + AS T
Sbjct: 326 SSPSTSSSTTLAPKSSTR*PTEPPSSPPCSRPPAPPPEPPATSTSPTSKEAK*ASPLRTT 505
Query: 71 DMS 73
S
Sbjct: 506 TAS 514
Score = 31.6 bits (70), Expect = 0.26
Identities = 14/35 (40%), Positives = 19/35 (54%)
Frame = +2
Query: 33 PPSPPPVTVVEDPPSIHNAITTSPPSSSSSSASTS 67
PP PPP P+ TSPPS+++S + TS
Sbjct: 116 PPQPPPPMAATTLPAFSPNTPTSPPSTTTSPSPTS 220
Score = 30.4 bits (67), Expect = 0.58
Identities = 22/63 (34%), Positives = 31/63 (48%)
Frame = +2
Query: 7 PDWLNSPMWSSPTEQNRFSATSYPSEPPSPPPVTVVEDPPSIHNAITTSPPSSSSSSAST 66
P SP +SP +A PS P + PP PPS + + SPP +SS +++
Sbjct: 188 PSTTTSPSPTSPARST--TAKPSPSSPSTTPPC-----PPS-STSTSPSPP*KTSSPSTS 343
Query: 67 SST 69
SST
Sbjct: 344 SST 352
>TC13980 similar to UP|Q9SGT0 (Q9SGT0) T6H22.18 protein (F14J16.33), partial
(12%)
Length = 565
Score = 38.1 bits (87), Expect = 0.003
Identities = 24/73 (32%), Positives = 32/73 (42%), Gaps = 8/73 (10%)
Frame = +2
Query: 5 RVPDWLNSPMWSSPTEQNRFSATS--------YPSEPPSPPPVTVVEDPPSIHNAITTSP 56
+ P + P SSPT + S ++ P PP PPP PS H + +P
Sbjct: 320 KTPSPCSPPTSSSPTGSSSHSTSNPPHPPPQQQPFSPPPPPPPPPSPTAPSSHPRLPAAP 499
Query: 57 PSSSSSSASTSST 69
+ SSAS SST
Sbjct: 500 AAGRISSASKSST 538
>TC14641 similar to UP|NTF2_ARATH (Q9C7F5) Nuclear transport factor 2
(NTF-2), partial (98%)
Length = 760
Score = 38.1 bits (87), Expect = 0.003
Identities = 26/81 (32%), Positives = 38/81 (46%), Gaps = 4/81 (4%)
Frame = +2
Query: 6 VPDWLNS----PMWSSPTEQNRFSATSYPSEPPSPPPVTVVEDPPSIHNAITTSPPSSSS 61
VPDW S P S +++ TS PS PP P PP +T SPP+S++
Sbjct: 98 VPDWRISTRKVPC*PSRVRRSKAPPTSSPSSPPFPSSSASTPSPP-----LTASPPASTT 262
Query: 62 SSASTSSTDDMSISRLAQSQS 82
+ +S+S+ S + S S
Sbjct: 263 ACSSSSAVISNSPASSTPSSS 325
>AV428818
Length = 417
Score = 37.7 bits (86), Expect = 0.004
Identities = 21/51 (41%), Positives = 28/51 (54%)
Frame = +3
Query: 18 PTEQNRFSATSYPSEPPSPPPVTVVEDPPSIHNAITTSPPSSSSSSASTSS 68
P ++T+ PS PSP T PPS N+ TSPPS+ +S+ S SS
Sbjct: 207 PLSPRTSTSTASPSPSPSPNSKTATTSPPST-NSSGTSPPSALTSATSVSS 356
Score = 29.6 bits (65), Expect = 0.99
Identities = 25/79 (31%), Positives = 35/79 (43%), Gaps = 16/79 (20%)
Frame = +3
Query: 6 VPDWLNSPMWSSPTEQNRFSATSYPSEPPSPP--------------PVTVVEDPPSIHNA 51
+P LN ++ SP + SAT + PPS P P + +P NA
Sbjct: 12 LPKTLN--LFPSPPQWRPVSATLSAATPPSSPQTHQRH*PQRRSQEPPSTSSNPKPTRNA 185
Query: 52 ITTS--PPSSSSSSASTSS 68
+TS PP S +S ST+S
Sbjct: 186SSTSAAPPLSPRTSTSTAS 242
>TC11009 similar to GB|AAP21377.1|30102918|BT006569 At1g47970 {Arabidopsis
thaliana;}, partial (44%)
Length = 700
Score = 37.4 bits (85), Expect = 0.005
Identities = 24/67 (35%), Positives = 36/67 (52%), Gaps = 4/67 (5%)
Frame = -2
Query: 13 PMWSSPTEQNRFSATSYPSEPPSPPPVTVVEDPPSIHNAITTS----PPSSSSSSASTSS 68
P SS + + S++S S SPPP PPS ++ ++S PP +SSSS+S+ S
Sbjct: 237 PKSSSSSSSSSSSSSSSSSSSGSPPPPRPPSTPPSSSSSSSSSSSSPPPGASSSSSSSPS 58
Query: 69 TDDMSIS 75
+ S S
Sbjct: 57 SSSPSSS 37
Score = 27.7 bits (60), Expect = 3.7
Identities = 20/57 (35%), Positives = 28/57 (49%)
Frame = -2
Query: 12 SPMWSSPTEQNRFSATSYPSEPPSPPPVTVVEDPPSIHNAITTSPPSSSSSSASTSS 68
SP P S++S S SP PP ++ ++SP SSS SS+S+SS
Sbjct: 171 SPPPPRPPSTPPSSSSSSSSSSSSP--------PPGASSSSSSSPSSSSPSSSSSSS 25
>TC11881 similar to PIR|T48303|T48303 meiosis specific-like protein -
Arabidopsis thaliana {Arabidopsis thaliana;}, partial
(34%)
Length = 631
Score = 37.0 bits (84), Expect = 0.006
Identities = 27/70 (38%), Positives = 37/70 (52%)
Frame = +3
Query: 12 SPMWSSPTEQNRFSATSYPSEPPSPPPVTVVEDPPSIHNAITTSPPSSSSSSASTSSTDD 71
SP PT ++T+ S +PP +V PPS A SPP+SSSSS S++S
Sbjct: 360 SPPAPPPTSSMSLNSTASSSRI-NPPSARLVTSPPS---ANPPSPPASSSSSTSSASRGF 527
Query: 72 MSISRLAQSQ 81
MS S + +Q
Sbjct: 528 MSPSVTSSTQ 557
Score = 35.0 bits (79), Expect = 0.023
Identities = 31/101 (30%), Positives = 48/101 (46%), Gaps = 16/101 (15%)
Frame = +3
Query: 2 AKKRVPDWLNSPMWSSPTEQNRFSATSYPSEPPSPPPVTVVE----------DPPSIHNA 51
A+ R P +SP+ +SP+ PP+PPP + + +PPS
Sbjct: 312 ARSRTP---SSPVTASPS-----------MSPPAPPPTSSMSLNSTASSSRINPPSAR-- 443
Query: 52 ITTSPPSSS------SSSASTSSTDDMSISRLAQSQSQLLA 86
+ TSPPS++ SSS+STSS +S S +Q+L+
Sbjct: 444 LVTSPPSANPPSPPASSSSSTSSASRGFMSPSVTSSTQMLS 566
>TC17283 similar to UP|O65760 (O65760) Extensin (Fragment), partial (16%)
Length = 513
Score = 36.6 bits (83), Expect = 0.008
Identities = 18/43 (41%), Positives = 26/43 (59%), Gaps = 5/43 (11%)
Frame = +1
Query: 30 PSEPPSPP-----PVTVVEDPPSIHNAITTSPPSSSSSSASTS 67
PS PP PP P T+ PP+ + +TT PP +SSS+ S++
Sbjct: 88 PSPPPPPPTTKHSPETLQAPPPATPSTLTTHPPPTSSSAFSST 216
Score = 28.5 bits (62), Expect = 2.2
Identities = 16/54 (29%), Positives = 25/54 (45%), Gaps = 4/54 (7%)
Frame = +1
Query: 18 PTEQNRFSAT----SYPSEPPSPPPVTVVEDPPSIHNAITTSPPSSSSSSASTS 67
PT + FS+T + PPS P + P A TSPP ++ +++S
Sbjct: 187 PTSSSAFSSTLPIPRWNPPPPSRTPTSTYTSTPPFSAAPNTSPPYCPTAGSASS 348
>TC10688 similar to UP|AAR10812 (AAR10812) Superoxide dismutase, partial
(91%)
Length = 822
Score = 36.6 bits (83), Expect = 0.008
Identities = 20/58 (34%), Positives = 28/58 (47%)
Frame = +2
Query: 17 SPTEQNRFSATSYPSEPPSPPPVTVVEDPPSIHNAITTSPPSSSSSSASTSSTDDMSI 74
SP + TS PS PP+P P + PP + + +PP S +SS S T D +
Sbjct: 110 SPVPPSNSPPTSEPSHPPNPSPSS---PPPRKPSPSSRAPPPSKASSPSPKKTTDQQL 274
>TC11354 similar to UP|CAE54480 (CAE54480) Alpha glucosidase II , partial
(6%)
Length = 583
Score = 36.6 bits (83), Expect = 0.008
Identities = 22/65 (33%), Positives = 33/65 (49%)
Frame = -1
Query: 4 KRVPDWLNSPMWSSPTEQNRFSATSYPSEPPSPPPVTVVEDPPSIHNAITTSPPSSSSSS 63
K +P P SPT + S SPP ++ PPS N+ T +PPSS+++
Sbjct: 193 KTLPSTRPRPASESPTSSSPSSPPPSSGSSVSPPRISPPPPPPS--NSPTATPPSSATNP 20
Query: 64 ASTSS 68
+S+SS
Sbjct: 19 SSSSS 5
Score = 31.2 bits (69), Expect = 0.34
Identities = 19/54 (35%), Positives = 25/54 (46%)
Frame = -1
Query: 16 SSPTEQNRFSATSYPSEPPSPPPVTVVEDPPSIHNAITTSPPSSSSSSASTSST 69
SS T SA+ PP PPP PP + TSPP+SS ++ + T
Sbjct: 406 SSETAIRPHSASEPDPAPPPPPP----SSPPMSPSPTATSPPTSSPNTHPPTPT 257
Score = 31.2 bits (69), Expect = 0.34
Identities = 24/70 (34%), Positives = 32/70 (45%)
Frame = -1
Query: 13 PMWSSPTEQNRFSATSYPSEPPSPPPVTVVEDPPSIHNAITTSPPSSSSSSASTSSTDDM 72
PM SPT ATS P+ P+ P T +P S + TT+ S+S S+ + ST
Sbjct: 328 PMSPSPT------ATSPPTSSPNTHPPTPTPNP*SSPSPSTTTASSASPSTKTLPSTRPR 167
Query: 73 SISRLAQSQS 82
S S S
Sbjct: 166 PASESPTSSS 137
>BP067133
Length = 507
Score = 36.2 bits (82), Expect = 0.011
Identities = 24/63 (38%), Positives = 33/63 (52%)
Frame = +1
Query: 16 SSPTEQNRFSATSYPSEPPSPPPVTVVEDPPSIHNAITTSPPSSSSSSASTSSTDDMSIS 75
SSPT + + S PPS P T PPSI + T +P +SS S ++ST S +
Sbjct: 325 SSPTASPSSPSPTPASSPPSAPSRT---SPPSIPFSNTWTPSASSLISGPSTSTSHYSAA 495
Query: 76 RLA 78
R+A
Sbjct: 496 RIA 504
>TC16838 similar to UP|BAD07764 (BAD07764) GTPase activating protein-like,
partial (16%)
Length = 586
Score = 36.2 bits (82), Expect = 0.011
Identities = 11/39 (28%), Positives = 24/39 (61%)
Frame = +1
Query: 374 FYAFRWITLLLTQEFDFADSLRIWDTLVSDPDGPQVQIY 412
F+ FRW+ + +EF++ D++R+W+ L + + +Y
Sbjct: 19 FFCFRWVLIPFKREFEYEDTMRLWEVLWTHYPSEHLHLY 135
>TC13755 similar to UP|Q8HCN7 (Q8HCN7) Ribosomal protein L2, partial (14%)
Length = 469
Score = 36.2 bits (82), Expect = 0.011
Identities = 26/65 (40%), Positives = 32/65 (49%), Gaps = 14/65 (21%)
Frame = +2
Query: 30 PSEPPSPP---------PVTVVEDP-----PSIHNAITTSPPSSSSSSASTSSTDDMSIS 75
PSEPP PP P T +P PSI N T PPS+SS +ST+ TD ++
Sbjct: 35 PSEPPRPPAETPKAA*LPSTEAAEPSACNEPSISNE-TPLPPSASSRGSSTTLTDPLTSP 211
Query: 76 RLAQS 80
A S
Sbjct: 212SFAGS 226
>TC18261 similar to UP|WSC2_YEAST (P53832) Cell wall integrity and stress
response component 2 precursor, partial (5%)
Length = 538
Score = 36.2 bits (82), Expect = 0.011
Identities = 27/74 (36%), Positives = 38/74 (50%)
Frame = +3
Query: 16 SSPTEQNRFSATSYPSEPPSPPPVTVVEDPPSIHNAITTSPPSSSSSSASTSSTDDMSIS 75
S P N +A S P+ PP PPP+ PP+ + +T++ +SSSS S SS S S
Sbjct: 162 SKPNPLN--AAQSPPTSPPQPPPL----QPPTSLSLMTSTTSTSSSSPNSRSSA--ASAS 317
Query: 76 RLAQSQSQLLAEVP 89
+ S S + VP
Sbjct: 318 SSSPSPSSVKTTVP 359
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.327 0.139 0.431
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,357,302
Number of Sequences: 28460
Number of extensions: 140947
Number of successful extensions: 5084
Number of sequences better than 10.0: 524
Number of HSP's better than 10.0 without gapping: 2942
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 4147
length of query: 424
length of database: 4,897,600
effective HSP length: 93
effective length of query: 331
effective length of database: 2,250,820
effective search space: 745021420
effective search space used: 745021420
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 56 (26.2 bits)
Medicago: description of AC146683.4


