

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146651.1 - phase: 0
(935 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
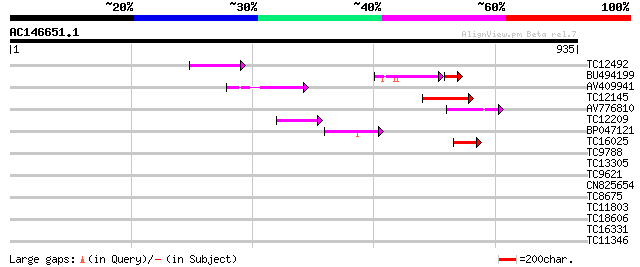
Score E
Sequences producing significant alignments: (bits) Value
TC12492 weakly similar to UP|AAS53933 (AAS53933) AFR562Cp, parti... 82 4e-16
BU494199 63 1e-14
AV409941 72 5e-13
TC12145 similar to UP|Q9ZVP6 (Q9ZVP6) T7A14.1 protein (Fragment)... 71 9e-13
AV776810 59 4e-09
TC12209 similar to UP|O60177 (O60177) DEAD box helicase, partial... 52 4e-07
BP047121 48 6e-06
TC16025 weakly similar to UP|SN21_HUMAN (P28370) Possible global... 42 4e-04
TC9788 similar to PIR|T47605|T47605 RING finger-like protein - A... 40 0.002
TC13305 weakly similar to UP|O29528 (O29528) Cobalt transport pr... 35 0.071
TC9621 similar to UP|Q9SIW2 (Q9SIW2) At2g16390 protein, partial ... 34 0.12
CN825654 29 3.0
TC8675 weakly similar to UP|Q9LFH0 (Q9LFH0) PDR9 ABC transporter... 29 3.0
TC11803 similar to UP|Q9ZVP6 (Q9ZVP6) T7A14.1 protein (Fragment)... 28 5.1
TC18606 similar to UP|AAR89453 (AAR89453) Zeta-sarcoglycan, part... 28 6.7
TC16331 similar to GB|AAN64534.1|24797044|BT001143 At5g12410/At5... 28 6.7
TC11346 similar to GB|AAD18096.1|4337175|T31J12 ESTs gb|T20589, ... 28 6.7
>TC12492 weakly similar to UP|AAS53933 (AAS53933) AFR562Cp, partial (7%)
Length = 922
Score = 82.0 bits (201), Expect = 4e-16
Identities = 39/93 (41%), Positives = 55/93 (58%)
Frame = +1
Query: 297 EQPKELRGGSLFPHQLEALNWLRKCWYKSRNVILADEMGLGKTISACAFISSLYFEFKVS 356
EQP L+GG L +Q+E L W+ + + N ILADEMGLGKTI + I+ L V+
Sbjct: 628 EQPSILQGGELRSYQIEGLQWMLSLFNNNLNGILADEMGLGKTIQTISLIAHLMEYKGVT 807
Query: 357 RPCLVLVPLVTMGNWLAEFALWAPDVNVVQYHG 389
P L++ P + NW+ EF+ WAP + + Y G
Sbjct: 808 GPHLIVAPKAVLPNWMNEFSTWAPSIKTILYDG 906
>BU494199
Length = 570
Score = 63.2 bits (152), Expect(2) = 1e-14
Identities = 52/141 (36%), Positives = 66/141 (45%), Gaps = 27/141 (19%)
Frame = +1
Query: 602 GTEPDSGSVEF-------------LHEMRIKASAKLTLLHSMLK-------ILYKEG--- 638
GT PDS S F LH++ S KL LH +L+ EG
Sbjct: 7 GTSPDSFSAIFSELFPAGSDVISELHKLH--HSPKLVALHEILEECGIGVDASGSEGTVS 180
Query: 639 ---HRVLIFSQMTKLLDILE-DYLNIEFGPKTYERVDGSVSVTDRQTAIARFNQDKSRFV 694
HRVLIF+Q LDI+E D TY R+DGSV R + FN D + V
Sbjct: 181 IGQHRVLIFAQHKAFLDIIERDLFQTHMKNVTYLRLDGSVEPEKRFEIVKAFNSDPTIDV 360
Query: 695 FLLSTRSCGLGINLATADTVI 715
LL+T GLG+NL +ADT++
Sbjct: 361 LLLTTHVGGLGLNLTSADTLV 423
Score = 34.3 bits (77), Expect(2) = 1e-14
Identities = 15/29 (51%), Positives = 21/29 (71%)
Frame = +2
Query: 718 DSDFNPHADIQAMNRAHRIGQSNRLLVYR 746
+ D+NP D QAM+RAHR+GQ + V+R
Sbjct: 431 EHDWNPMRDHQAMDRAHRLGQKKVVNVHR 517
>AV409941
Length = 429
Score = 71.6 bits (174), Expect = 5e-13
Identities = 42/136 (30%), Positives = 66/136 (47%)
Frame = +1
Query: 358 PCLVLVPLVTMGNWLAEFALWAPDVNVVQYHGCAKARAIIRQYEWHASDPSGLNKKTEAY 417
P L++ P + NW EF+ W+ + +V YHG + K +A
Sbjct: 67 PVLIICPSSVIHNWEDEFSKWS-NFSVSIYHGANRDLVY---------------DKLQAN 198
Query: 418 KFNVLLTSYEMVLADYSHFRGVPWEVLIVDEGHRLKNSESKLFSLLNSISFQHRVLLTGT 477
+L+TS++ F + W V+I+DE HRLKN SKL+ I R LTGT
Sbjct: 199 GVEILITSFDSYRIHGRSFLDINWNVVIIDEAHRLKNDRSKLYKACLEIKTLRRYGLTGT 378
Query: 478 PLQNNLGEMYNLLNFL 493
+QN + E++NL +++
Sbjct: 379 VMQNKIMELFNLFDWV 426
>TC12145 similar to UP|Q9ZVP6 (Q9ZVP6) T7A14.1 protein (Fragment), partial
(76%)
Length = 664
Score = 70.9 bits (172), Expect = 9e-13
Identities = 36/83 (43%), Positives = 52/83 (62%)
Frame = +2
Query: 682 AIARFNQDKSRFVFLLSTRSCGLGINLATADTVIIYDSDFNPHADIQAMNRAHRIGQSNR 741
AI RFN D FLLS ++ G+ +NL A V + + +NP A+ QA +R HRIGQ+
Sbjct: 11 AIKRFNDDPDCKFFLLSLKAAGVALNLTVASHVFLMEPWWNPDAEQQAQDRIHRIGQNKP 190
Query: 742 LLVYRLVVRASVEERILQLAKKK 764
+ V + + +VEERIL+L +KK
Sbjct: 191 IRVVKFITENTVEERILELQQKK 259
>AV776810
Length = 360
Score = 58.9 bits (141), Expect = 4e-09
Identities = 34/94 (36%), Positives = 50/94 (53%)
Frame = +2
Query: 721 FNPHADIQAMNRAHRIGQSNRLLVYRLVVRASVEERILQLAKKKLMLDQLFKGKSGSQKE 780
+NP D QA +R HRIGQ+ + +YRL+ +++EE IL+ A +K LD L G E
Sbjct: 2 WNPAMDQQAQDRCHRIGQTREVHIYRLISESTIEENILKKANQKRALDDLVIQSGGYNTE 181
Query: 781 VEDILKWGTEELFNDSCALNGKDTSENNNSNKDE 814
K E+F+ L+ K+T + N N E
Sbjct: 182 F--FKKLDPMEIFSGHRTLSIKNTPKEKNQNNGE 277
>TC12209 similar to UP|O60177 (O60177) DEAD box helicase, partial (6%)
Length = 550
Score = 52.0 bits (123), Expect = 4e-07
Identities = 24/75 (32%), Positives = 42/75 (56%)
Frame = +1
Query: 441 WEVLIVDEGHRLKNSESKLFSLLNSISFQHRVLLTGTPLQNNLGEMYNLLNFLQPASFPS 500
W +I+DE +KN + ++ +R LTGTP+ NN+GE+Y+L++FL+ +
Sbjct: 277 WYRIILDEAQCIKNKSTGAARACCTLQAIYRFCLTGTPMMNNVGELYSLIHFLRIKPYNE 456
Query: 501 LSAFEERFNDLTSAE 515
F++ F LT +
Sbjct: 457 WKRFQDSFGCLTKPD 501
>BP047121
Length = 525
Score = 48.1 bits (113), Expect = 6e-06
Identities = 27/109 (24%), Positives = 52/109 (46%), Gaps = 12/109 (11%)
Frame = -1
Query: 520 LKKLVSPHMLRRLKKDAMQNIPPKTERMVPVELSSIQAEYYRAMLTKNYQILR------- 572
+K ++ P +LRRLK D MQ + PK +++ V + Q Y+ + + + +
Sbjct: 522 MKSILGPFILRRLKSDVMQQLVPKIQQVEYVVMEKQQESAYKEAIEEYRAVSQARMAKCS 343
Query: 573 -----NIGKGIAQQSMLNIVMQLRKVCNHPYLIPGTEPDSGSVEFLHEM 616
N+ + + ++ + N +Q RK+ NHP LI D + F ++
Sbjct: 342 ELNSNNVLEVLPRRQINNYFVQFRKIANHPLLIRRIYSDEDVIRFARKL 196
>TC16025 weakly similar to UP|SN21_HUMAN (P28370) Possible global
transcription activator SNF2L1, partial (3%)
Length = 550
Score = 42.0 bits (97), Expect = 4e-04
Identities = 23/46 (50%), Positives = 29/46 (63%)
Frame = +3
Query: 732 RAHRIGQSNRLLVYRLVVRASVEERILQLAKKKLMLDQLFKGKSGS 777
RAHRIGQ+ + V RL VR +VE+RIL L KK + G+ GS
Sbjct: 3 RAHRIGQTRPVTVLRLTVRDTVEDRILALQPKKRKMVSSAFGEDGS 140
>TC9788 similar to PIR|T47605|T47605 RING finger-like protein - Arabidopsis
thaliana {Arabidopsis thaliana;}, partial (6%)
Length = 696
Score = 40.0 bits (92), Expect = 0.002
Identities = 23/77 (29%), Positives = 43/77 (54%)
Frame = +2
Query: 683 IARFNQDKSRFVFLLSTRSCGLGINLATADTVIIYDSDFNPHADIQAMNRAHRIGQSNRL 742
+A F D S V L+ S L ++L+ V + + ++ + Q ++RAHR+G S +
Sbjct: 2 LAMFQHDSSCMVLLMDG-SAALVLDLSFVSHVFLMEPIWDRSLEEQVISRAHRMGASRPI 178
Query: 743 LVYRLVVRASVEERILQ 759
V L +R ++EE++L+
Sbjct: 179 HVETLAMRGTIEEQMLE 229
>TC13305 weakly similar to UP|O29528 (O29528) Cobalt transport protein
(CBIQ-1), partial (8%)
Length = 488
Score = 34.7 bits (78), Expect = 0.071
Identities = 15/54 (27%), Positives = 31/54 (56%)
Frame = +2
Query: 707 NLATADTVIIYDSDFNPHADIQAMNRAHRIGQSNRLLVYRLVVRASVEERILQL 760
+L A + + + +P + + ++ HRIG + V +L++ S+EE+IL+L
Sbjct: 35 DLTAASRIYLMEPYLSPEIEARMIDLVHRIGPDQIVNVVKLIMHNSIEEKILEL 196
>TC9621 similar to UP|Q9SIW2 (Q9SIW2) At2g16390 protein, partial (10%)
Length = 588
Score = 33.9 bits (76), Expect = 0.12
Identities = 26/77 (33%), Positives = 37/77 (47%), Gaps = 6/77 (7%)
Frame = +1
Query: 722 NPHADIQAMNRAHRIGQSNRLLVYRLVVRASVEE-RILQLAKKKLMLDQLFK-----GKS 775
NP QA+ RA R GQ+ ++ VYRL+ S EE + KK+L+ F+ G
Sbjct: 7 NPSVTRQAIGRAFRPGQTKKVFVYRLIAADSPEEDDHITCFKKELISKMWFEWNEYCGDR 186
Query: 776 GSQKEVEDILKWGTEEL 792
Q E + + G E L
Sbjct: 187 AFQVETVSVNECGDEFL 237
>CN825654
Length = 580
Score = 29.3 bits (64), Expect = 3.0
Identities = 24/80 (30%), Positives = 41/80 (51%), Gaps = 2/80 (2%)
Frame = +2
Query: 13 SACDNDLDGEIAENLVHDPQNVKSSDEGELHNTDRVEKIHVY--RRSITKESKNGNLLNS 70
+A D+D DG++ +N S D+G N D E + + ++ +K SK G L S
Sbjct: 344 AAFDDDNDGDVMDN---------SVDDGRDGNDDVDEPVIAFTGKKKSSKGSKKGGALFS 496
Query: 71 LSKATDDLGSCARDGTDQDD 90
+ + DD+ A+D ++DD
Sbjct: 497 -AASFDDVDE-AKDDKEEDD 550
>TC8675 weakly similar to UP|Q9LFH0 (Q9LFH0) PDR9 ABC transporter, partial
(13%)
Length = 727
Score = 29.3 bits (64), Expect = 3.0
Identities = 13/43 (30%), Positives = 20/43 (46%)
Frame = +3
Query: 360 LVLVPLVTMGNWLAEFALWAPDVNVVQYHGCAKARAIIRQYEW 402
++ +PLV W A+F +W V GC + I R+ W
Sbjct: 360 VLFMPLVLGNEWDADFTIWRYKQKYVSL*GCQNSCYIFRRLLW 488
>TC11803 similar to UP|Q9ZVP6 (Q9ZVP6) T7A14.1 protein (Fragment), partial
(56%)
Length = 519
Score = 28.5 bits (62), Expect = 5.1
Identities = 14/37 (37%), Positives = 23/37 (61%)
Frame = +1
Query: 744 VYRLVVRASVEERILQLAKKKLMLDQLFKGKSGSQKE 780
+ R V+ ++EERIL+L +KK + +F+G G E
Sbjct: 13 IVRFVIENTIEERILKLQEKK---ELVFEGTVGGSSE 114
>TC18606 similar to UP|AAR89453 (AAR89453) Zeta-sarcoglycan, partial (6%)
Length = 319
Score = 28.1 bits (61), Expect = 6.7
Identities = 10/24 (41%), Positives = 14/24 (57%)
Frame = +3
Query: 160 EEVSYEFLVKWVGKSHIHNSWISE 183
EE S+ F+ W GK+ N W+ E
Sbjct: 51 EEESFSFVFVWDGKNEEENEWVGE 122
>TC16331 similar to GB|AAN64534.1|24797044|BT001143 At5g12410/At5g12410
{Arabidopsis thaliana;}, partial (42%)
Length = 932
Score = 28.1 bits (61), Expect = 6.7
Identities = 20/65 (30%), Positives = 30/65 (45%), Gaps = 2/65 (3%)
Frame = +2
Query: 856 LDRSNLQDAST-DIAEGDSEN-DMLGSMKALEWNDEPTEEHVEGESPPHGADDMCTQNSE 913
LD S +DA + AEGD N D + M + + + + G+ PH DD+
Sbjct: 158 LDSSVDEDAGIGEKAEGDEANGDKMPKMDSSVNENAGHDNEIGGKENPHKIDDVHAAAGN 337
Query: 914 KKEDN 918
K +DN
Sbjct: 338 KTDDN 352
>TC11346 similar to GB|AAD18096.1|4337175|T31J12 ESTs gb|T20589, gb|T04648,
gb|AA597906, gb|T04111, gb|R84180, gb|R65428, gb|T44439,
gb|T76570, gb|R90004, gb|T45020, gb|T42457, gb|T20921,
gb|AA042762 and gb|AA720210 come from this gene.
{Arabidopsis thaliana;}, partial (79%)
Length = 555
Score = 28.1 bits (61), Expect = 6.7
Identities = 26/104 (25%), Positives = 44/104 (42%)
Frame = +2
Query: 132 SLETKQKEVVLEKGMGSSGDNKVQDSIGEEVSYEFLVKWVGKSHIHNSWISESHLKVIAK 191
SLE+KQ+E +++ SS + + EE+ G+S I++ + +V +K
Sbjct: 29 SLESKQRESEIDRSSSSSSSSSSMSLVTEEIR--------GRSEIYH---GDELCQVKSK 175
Query: 192 RKLENYKAKYGTATINICEEQWKNPERLLAIRTSKQGTSEAFVK 235
L+ G + EE N E L K+ T+ F K
Sbjct: 176 DLLKEIALPNGLLPLKDMEECGINRESGLVWLKQKKSTTHKFDK 307
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.314 0.131 0.378
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,964,303
Number of Sequences: 28460
Number of extensions: 192961
Number of successful extensions: 789
Number of sequences better than 10.0: 34
Number of HSP's better than 10.0 without gapping: 785
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 788
length of query: 935
length of database: 4,897,600
effective HSP length: 99
effective length of query: 836
effective length of database: 2,080,060
effective search space: 1738930160
effective search space used: 1738930160
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 60 (27.7 bits)
Medicago: description of AC146651.1


