

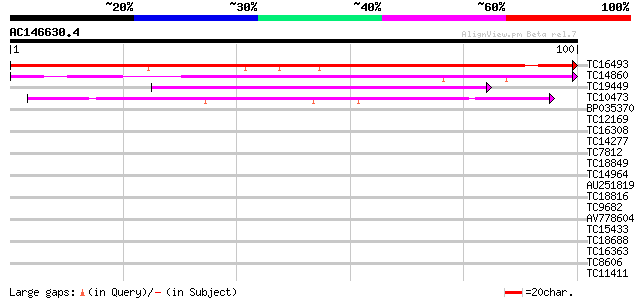
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146630.4 + phase: 0
(100 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC16493 similar to UP|Q7XTG2 (Q7XTG2) OJ991214_12.5 protein, par... 142 9e-36
TC14860 similar to UP|TPIS_COPJA (P21820) Triosephosphate isomer... 75 2e-15
TC19449 similar to PIR|E86300|E86300 protein F3O9.30 [imported] ... 52 2e-08
TC10473 similar to UP|Q7XBE3 (Q7XBE3) Humj1, partial (69%) 42 1e-05
BP035370 33 0.006
TC12169 homologue to UP|TF2B_SOYBN (P48513) Transcription initia... 31 0.030
TC16308 similar to GB|AAM16172.1|20334832|AY094016 At2g39720/T5I... 30 0.051
TC14277 similar to UP|Q9SHY3 (Q9SHY3) F1E22.9 (AT1G65720/F1E22_1... 29 0.11
TC7812 UP|Q9LKJ6 (Q9LKJ6) Water-selective transport intrinsic me... 28 0.19
TC18849 28 0.25
TC14964 weakly similar to UP|SOX_ARATH (Q9SJA7) Potential sarcos... 28 0.25
AU251819 28 0.25
TC18816 28 0.25
TC9682 similar to UP|Q8VZC0 (Q8VZC0) DTDP-glucose 4-6-dehydratas... 28 0.33
AV778604 28 0.33
TC15433 weakly similar to UP|Q9FEQ0 (Q9FEQ0) Glycerol-3-phosphat... 28 0.33
TC18688 weakly similar to UP|Q9LP77 (Q9LP77) T1N15.9, partial (13%) 27 0.43
TC16363 UP|Q84RQ3 (Q84RQ3) Sucrose transporter 4 protein, complete 27 0.43
TC8606 similar to UP|AAQ82688 (AAQ82688) Epa4p, partial (7%) 27 0.43
TC11411 similar to UP|Q8RY22 (Q8RY22) AT3g03380/T21P5_20, partia... 27 0.43
>TC16493 similar to UP|Q7XTG2 (Q7XTG2) OJ991214_12.5 protein, partial (33%)
Length = 769
Score = 142 bits (358), Expect = 9e-36
Identities = 81/115 (70%), Positives = 91/115 (78%), Gaps = 15/115 (13%)
Frame = +2
Query: 1 MSRRNGSGPKLDLKLNLSPPRVN-RRMESSPTRSASVSPPS---SCVSSE---------- 46
MS RNGSGPK +LKLNLSPPRVN RR+ESSP RSA+VSP S SCVS+E
Sbjct: 5 MSHRNGSGPKPELKLNLSPPRVNHRRLESSPNRSATVSPTSPTTSCVSTELNQEEGNNNH 184
Query: 47 NGSSSPE-ATSMLLVGCPRCLMYVMLSENDPKCPKCHSTVLLDFLNAENNNNKKS 100
N S+SPE ATSM+LVGCPRCLMYVMLSE+DPKCPKC STVLLDFL+ +NN +KS
Sbjct: 185 NHSTSPEEATSMVLVGCPRCLMYVMLSEDDPKCPKCKSTVLLDFLH--DNNKRKS 343
>TC14860 similar to UP|TPIS_COPJA (P21820) Triosephosphate isomerase,
cytosolic (TIM) , partial (68%)
Length = 917
Score = 74.7 bits (182), Expect = 2e-15
Identities = 46/104 (44%), Positives = 55/104 (52%), Gaps = 4/104 (3%)
Frame = +1
Query: 1 MSRRNGSGPKLDLKLNLSPPRVNRRMESSPTRSASVSPPSSCVSSENGSSSPEATSMLLV 60
MS R G L+ +L+LSPP ++A+ S SCVS+ G M L
Sbjct: 76 MSERAG----LEFELSLSPP----------LQAANSSSEGSCVSNSEGDDGETTKDMFLA 213
Query: 61 GCPRCLMYVMLSEND--PKCPKCHSTVL--LDFLNAENNNNKKS 100
GC CLMYV LS D PKCPKC STVL +DFLN NNN +
Sbjct: 214 GCLHCLMYVFLSSADPNPKCPKCKSTVLDIIDFLNNNQNNNNNN 345
>TC19449 similar to PIR|E86300|E86300 protein F3O9.30 [imported] -
Arabidopsis thaliana {Arabidopsis thaliana;}, partial
(24%)
Length = 600
Score = 51.6 bits (122), Expect = 2e-08
Identities = 23/60 (38%), Positives = 35/60 (58%)
Frame = +1
Query: 26 MESSPTRSASVSPPSSCVSSENGSSSPEATSMLLVGCPRCLMYVMLSENDPKCPKCHSTV 85
+ SSP+ S S S S +E S + GCP CL YV++++N+PKCP+C++ V
Sbjct: 166 LSSSPSASFSSSSSSIITQAEEECEVKILASPVATGCPGCLTYVLVTKNNPKCPRCNTNV 345
>TC10473 similar to UP|Q7XBE3 (Q7XBE3) Humj1, partial (69%)
Length = 786
Score = 42.4 bits (98), Expect = 1e-05
Identities = 36/101 (35%), Positives = 47/101 (45%), Gaps = 8/101 (7%)
Frame = +3
Query: 4 RNGSGPKLDLKLNLSPPRVNRRMESSPTRS---ASVSPPSSCVSSENGSSSP----EATS 56
+NG K D + S VN R S S + S SC SS +P E +
Sbjct: 246 KNGMKAKEDPR-RTSEYNVNNRYYESEEESRYDSEESSSESCTSSSKEHYAPLSCVEKQN 422
Query: 57 MLLV-GCPRCLMYVMLSENDPKCPKCHSTVLLDFLNAENNN 96
+L+V GC CLMY M+ + CPKC S LL F +EN +
Sbjct: 423 VLVVAGCKSCLMYFMVPKQVEDCPKC-SGQLLHFDRSENGS 542
>BP035370
Length = 563
Score = 33.5 bits (75), Expect = 0.006
Identities = 16/52 (30%), Positives = 25/52 (47%), Gaps = 4/52 (7%)
Frame = -2
Query: 34 ASVSPPSSCVSSEN----GSSSPEATSMLLVGCPRCLMYVMLSENDPKCPKC 81
+S + SCV ++ S + M+ C RC + VML ++ P CP C
Sbjct: 490 SSNNKKDSCVLTQTPSWLSSEGDDHNEMIAAVCMRCHLLVMLCKSSPSCPNC 335
>TC12169 homologue to UP|TF2B_SOYBN (P48513) Transcription initiation factor
IIB (General transcription factor TFIIB), partial (32%)
Length = 351
Score = 31.2 bits (69), Expect = 0.030
Identities = 19/51 (37%), Positives = 26/51 (50%), Gaps = 1/51 (1%)
Frame = -2
Query: 4 RNGSGPKLDLKLNLS-PPRVNRRMESSPTRSASVSPPSSCVSSENGSSSPE 53
R P L L++ + PP V R +E PTR+ S+SP S + SS E
Sbjct: 308 RKSPEPPLGLEIMVERPPSVRRGLEGPPTRTGSLSPDSLAKVRHSEVSSME 156
>TC16308 similar to GB|AAM16172.1|20334832|AY094016 At2g39720/T5I7.2
{Arabidopsis thaliana;}, partial (13%)
Length = 640
Score = 30.4 bits (67), Expect = 0.051
Identities = 20/57 (35%), Positives = 26/57 (45%), Gaps = 1/57 (1%)
Frame = +3
Query: 28 SSPTRSASVSPPSSCVSSENGSSSPEATSMLLVGCPRCLMYV-MLSENDPKCPKCHS 83
S+PTRS + P + N + P TS C C +V +L ND CP C S
Sbjct: 27 SNPTRSTQLVP-----NKMNSDTQPATTSSYW--CYSCNRFVHLLDHNDVVCPHCQS 176
>TC14277 similar to UP|Q9SHY3 (Q9SHY3) F1E22.9 (AT1G65720/F1E22_13), partial
(33%)
Length = 941
Score = 29.3 bits (64), Expect = 0.11
Identities = 23/58 (39%), Positives = 27/58 (45%), Gaps = 7/58 (12%)
Frame = +3
Query: 6 GSGPKLDLKLNLSPPRVNR-------RMESSPTRSASVSPPSSCVSSENGSSSPEATS 56
GS P L SPP V R+ SS A++SPP C SS G SSP T+
Sbjct: 345 GSPPLLT---TTSPPSVTAPRTSSASRLPSSSALDAALSPPLPCTSS--GPSSPPVTT 503
Score = 24.3 bits (51), Expect = 3.6
Identities = 12/38 (31%), Positives = 16/38 (41%)
Frame = +3
Query: 19 PPRVNRRMESSPTRSASVSPPSSCVSSENGSSSPEATS 56
PP SSP ++ PP+ S SP +TS
Sbjct: 135 PPHAPAAASSSPPTPSATLPPTPSPPSPRSDPSPLSTS 248
Score = 23.5 bits (49), Expect = 6.2
Identities = 14/36 (38%), Positives = 17/36 (46%)
Frame = +3
Query: 28 SSPTRSASVSPPSSCVSSENGSSSPEATSMLLVGCP 63
S P+ S+S SPP + SSSP S L P
Sbjct: 96 SPPSSSSSPSPPPPPHAPAAASSSPPTPSATLPPTP 203
>TC7812 UP|Q9LKJ6 (Q9LKJ6) Water-selective transport intrinsic membrane
protein 1, complete
Length = 1149
Score = 28.5 bits (62), Expect = 0.19
Identities = 23/69 (33%), Positives = 29/69 (41%), Gaps = 4/69 (5%)
Frame = +1
Query: 19 PPRVNRRMESSPTRSASVSPPSSCVSSENGSS---SPEATSMLLVGCPRCLMYV-MLSEN 74
PP SPT S S SP S +S +S SP A+S + + LS +
Sbjct: 232 PPPPASSPPPSPTHSPSSSPSPSAPTSPAATSTPPSPSASSSAVTSPSSAVSSTSSLSFS 411
Query: 75 DPKCPKCHS 83
DP P C S
Sbjct: 412 DPSSPPCSS 438
>TC18849
Length = 770
Score = 28.1 bits (61), Expect = 0.25
Identities = 19/42 (45%), Positives = 25/42 (59%), Gaps = 3/42 (7%)
Frame = +1
Query: 18 SPPRVNRRMESSPTRSASVSPPS---SCVSSENGSSSPEATS 56
SPP + S+PT + + S PS S VSS++ SSS ATS
Sbjct: 184 SPPSITSPT-STPTPALTASSPSFTCSSVSSDSSSSSSTATS 306
>TC14964 weakly similar to UP|SOX_ARATH (Q9SJA7) Potential sarcosine oxidase
, partial (33%)
Length = 612
Score = 28.1 bits (61), Expect = 0.25
Identities = 13/40 (32%), Positives = 22/40 (54%)
Frame = +1
Query: 17 LSPPRVNRRMESSPTRSASVSPPSSCVSSENGSSSPEATS 56
++PP VN +SPT S + SP SS ++ P++ +
Sbjct: 151 VAPPTVNPAPSASPTPSTTTSPSSSPPTTYGNKPRPKSAT 270
Score = 24.6 bits (52), Expect = 2.8
Identities = 15/47 (31%), Positives = 22/47 (45%)
Frame = +1
Query: 15 LNLSPPRVNRRMESSPTRSASVSPPSSCVSSENGSSSPEATSMLLVG 61
L PP SS SA+ SP +S ++ + SSPE ++ G
Sbjct: 295 LTWPPPMTPSFTLSSRIASATTSPMNSSTAASSPRSSPENSTSRTTG 435
>AU251819
Length = 368
Score = 28.1 bits (61), Expect = 0.25
Identities = 22/53 (41%), Positives = 29/53 (54%), Gaps = 5/53 (9%)
Frame = +1
Query: 4 RNGSGPKLDLKLNLSPP--RVNRRMES---SPTRSASVSPPSSCVSSENGSSS 51
RNG+G LSPP ++N+ S SP+ S+S PPS SS + SSS
Sbjct: 88 RNGNG--------LSPPSLKINKDSHSIKKSPSSSSSPPPPSLSSSSSSISSS 222
>TC18816
Length = 465
Score = 28.1 bits (61), Expect = 0.25
Identities = 21/50 (42%), Positives = 24/50 (48%), Gaps = 1/50 (2%)
Frame = +3
Query: 3 RRNGSGPKLDLKLNLSPPRVNRRM-ESSPTRSASVSPPSSCVSSENGSSS 51
R SGP + KLN SP RV R E S RS S SCV G ++
Sbjct: 39 RGRESGPLANRKLNASPARVRRDAGEGSGRRSRS----PSCVRKVGGGAT 176
>TC9682 similar to UP|Q8VZC0 (Q8VZC0) DTDP-glucose 4-6-dehydratase-like
protein, partial (29%)
Length = 731
Score = 27.7 bits (60), Expect = 0.33
Identities = 17/49 (34%), Positives = 29/49 (58%), Gaps = 2/49 (4%)
Frame = +1
Query: 10 KLDLKLNLSPPRVNRRMESS--PTRSASVSPPSSCVSSENGSSSPEATS 56
K + KL+ +PP + S P+ ++SV+ SS SSE+ S+ P ++S
Sbjct: 238 KWEPKLHHTPPNPPNTPDPSPDPSTTSSVNSVSSSSSSESSSAPPSSSS 384
>AV778604
Length = 545
Score = 27.7 bits (60), Expect = 0.33
Identities = 18/45 (40%), Positives = 26/45 (57%), Gaps = 3/45 (6%)
Frame = -1
Query: 18 SPPRVNRRMESSPTRSASVSPPSSCVSSENGSSS---PEATSMLL 59
S + +R + S TRS SPPS+ ++S N SSS P+ +LL
Sbjct: 470 SRSKSSRWCKPSRTRSTMRSPPSTALASANPSSSSTPPQRLFLLL 336
>TC15433 weakly similar to UP|Q9FEQ0 (Q9FEQ0) Glycerol-3-phosphate
acyltransferase (Acyl-(acyl-carrier-protein):
glycerol-3-phosphate acyltransferase) (Fragment) ,
partial (6%)
Length = 640
Score = 27.7 bits (60), Expect = 0.33
Identities = 20/49 (40%), Positives = 24/49 (48%), Gaps = 5/49 (10%)
Frame = -1
Query: 13 LKLNLSPPRVNRRM-----ESSPTRSASVSPPSSCVSSENGSSSPEATS 56
L+L L P R NR + TR S PSS SS + +SSP A S
Sbjct: 214 LRLRLLPDRPNRTLPPPW*RRRLTRETPSSLPSSYPSSSSETSSPPAPS 68
>TC18688 weakly similar to UP|Q9LP77 (Q9LP77) T1N15.9, partial (13%)
Length = 514
Score = 27.3 bits (59), Expect = 0.43
Identities = 17/32 (53%), Positives = 18/32 (56%), Gaps = 1/32 (3%)
Frame = +3
Query: 30 PTRSASVSPPS-SCVSSENGSSSPEATSMLLV 60
PT S S SPPS S +S SS P TS L V
Sbjct: 249 PTSSRSASPPSPSPANSHMASSPPSPTSALSV 344
>TC16363 UP|Q84RQ3 (Q84RQ3) Sucrose transporter 4 protein, complete
Length = 2164
Score = 27.3 bits (59), Expect = 0.43
Identities = 16/40 (40%), Positives = 24/40 (60%)
Frame = +1
Query: 31 TRSASVSPPSSCVSSENGSSSPEATSMLLVGCPRCLMYVM 70
T AS PPS C+SS G S+ TS+ ++ P CL+ ++
Sbjct: 565 TPRASGRPPSPCLSSGFGFSTSLITSLKVLAEP-CLLILL 681
>TC8606 similar to UP|AAQ82688 (AAQ82688) Epa4p, partial (7%)
Length = 1008
Score = 27.3 bits (59), Expect = 0.43
Identities = 16/35 (45%), Positives = 19/35 (53%)
Frame = +1
Query: 18 SPPRVNRRMESSPTRSASVSPPSSCVSSENGSSSP 52
SPP + SS + S+S S PSS SS SS P
Sbjct: 346 SPPSSSSSSSSSSSSSSSWSSPSSSSSSSPFSSGP 450
Score = 26.9 bits (58), Expect = 0.56
Identities = 15/29 (51%), Positives = 18/29 (61%)
Frame = +1
Query: 28 SSPTRSASVSPPSSCVSSENGSSSPEATS 56
SS S+S SPPSS SS + SSS + S
Sbjct: 319 SSTPSSSSSSPPSSSSSSSSSSSSSSSWS 405
Score = 24.3 bits (51), Expect = 3.6
Identities = 17/48 (35%), Positives = 24/48 (49%), Gaps = 1/48 (2%)
Frame = +1
Query: 18 SPPRVNRRMESSPTRSASVSPPSS-CVSSENGSSSPEATSMLLVGCPR 64
SP + SSP+ S SP +S C+SS SP +ML+ P+
Sbjct: 556 SPNGPSPSSSSSPSSSLPSSPSASCCLSSL*DLVSPLVVAMLISNSPQ 699
>TC11411 similar to UP|Q8RY22 (Q8RY22) AT3g03380/T21P5_20, partial (19%)
Length = 728
Score = 27.3 bits (59), Expect = 0.43
Identities = 16/43 (37%), Positives = 25/43 (57%), Gaps = 3/43 (6%)
Frame = +3
Query: 19 PPRVN--RRMESSPTRSASVSPP-SSCVSSENGSSSPEATSML 58
PPR+ R ESSP S+S PP + + S ++P A+S++
Sbjct: 147 PPRIGGKRSTESSPPSSSSAPPPLAHSILSPPPPATPPASSLI 275
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.310 0.125 0.365
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,099,261
Number of Sequences: 28460
Number of extensions: 32016
Number of successful extensions: 445
Number of sequences better than 10.0: 179
Number of HSP's better than 10.0 without gapping: 401
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 433
length of query: 100
length of database: 4,897,600
effective HSP length: 76
effective length of query: 24
effective length of database: 2,734,640
effective search space: 65631360
effective search space used: 65631360
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 47 (22.7 bits)
Medicago: description of AC146630.4


