

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146630.11 + phase: 0
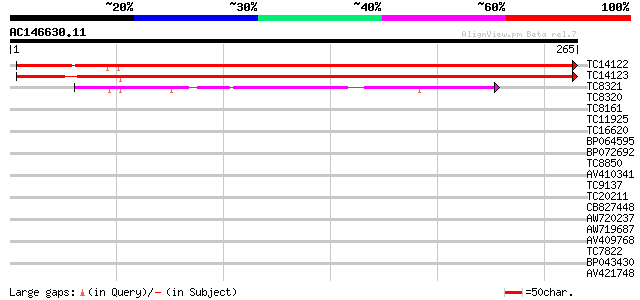
(265 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC14122 similar to UP|Q9FE12 (Q9FE12) Peroxiredoxin precursor, c... 451 e-128
TC14123 similar to UP|Q93X25 (Q93X25) 2-Cys peroxiredoxin, parti... 434 e-122
TC8321 similar to UP|AAS46230 (AAS46230) Peroxiredoxin Q, partia... 75 9e-15
TC8320 similar to UP|AAQ67661 (AAQ67661) Peroxiredoxin Q, partia... 37 0.005
TC8161 similar to UP|Q8LBV7 (Q8LBV7) Contains similarity to plas... 32 0.085
TC11925 similar to UP|Q9FKX6 (Q9FKX6) Prolyl 4-hydroxylase, alph... 30 0.42
TC16620 weakly similar to UP|Q9FN67 (Q9FN67) Gb|AAD56319.1, part... 29 0.94
BP064595 28 1.2
BP072692 28 1.6
TC8850 similar to UP|RK34_SPIOL (P82244) 50S ribosomal protein L... 28 1.6
AV410341 28 1.6
TC9137 UP|CAF02297 (CAF02297) Cysteine-rich polycomb-like protei... 28 1.6
TC20211 homologue to UP|Q08150 (Q08150) GTP-binding protein 6, p... 28 1.6
CB827448 28 1.6
AW720237 28 1.6
AW719687 28 2.1
AV409768 27 2.7
TC7822 similar to UP|Q42139 (Q42139) H+-transporting ATP synthas... 27 2.7
BP043430 27 3.6
AV421748 27 3.6
>TC14122 similar to UP|Q9FE12 (Q9FE12) Peroxiredoxin precursor, complete
Length = 1107
Score = 451 bits (1160), Expect = e-128
Identities = 236/265 (89%), Positives = 244/265 (92%), Gaps = 3/265 (1%)
Frame = +1
Query: 4 CSAPSASLLSSNPNILFSPKLSSPPHLSSLSIPNASNSLPK-LRTSL--PLSLNRFTSSR 60
CSAPSASLLSSNP ILF+ K SS + LS PN SNSL K LRTSL +S NR + S
Sbjct: 73 CSAPSASLLSSNPKILFTSKSSSSSS-APLSFPNFSNSLCKPLRTSLHPSISFNRSSRSP 249
Query: 61 RSFVVRASSELPLVGNAAPDFEAEAVFDQEFIKVKLSEYIGKKYVILFFYPLDFTFVCPT 120
RSFVVRASSELPLVGN APDFEAEAVFDQEFIKVKLSEYIGKKYVILFFYPLDFTFVCPT
Sbjct: 250 RSFVVRASSELPLVGNKAPDFEAEAVFDQEFIKVKLSEYIGKKYVILFFYPLDFTFVCPT 429
Query: 121 EITAFSDRHAEFAELNTEILGVSVDSVFSHLAWVQTDRKSGGLGDLNYPLVSDVTKSISK 180
EITAFSDRHAEF E+NTEILGVSVDSVFSHLAWVQT+RKSGGLGDLNYPLVSDVTKSISK
Sbjct: 430 EITAFSDRHAEFEEINTEILGVSVDSVFSHLAWVQTERKSGGLGDLNYPLVSDVTKSISK 609
Query: 181 SYGVLIPDQGIALRGLFIIDKEGVIQHSTINNLGIGRSVDETKRTLQALQYVQENPDEVC 240
SYGVLIPDQGIALRGLFIIDKEGVIQHSTINNLGIGRSVDETKRTLQALQYVQENPDEVC
Sbjct: 610 SYGVLIPDQGIALRGLFIIDKEGVIQHSTINNLGIGRSVDETKRTLQALQYVQENPDEVC 789
Query: 241 PAGWKPGEKSMKPDPKLSKEYFSAV 265
PAGWKPGEKSMKPDPKLSK+YF+AV
Sbjct: 790 PAGWKPGEKSMKPDPKLSKDYFAAV 864
>TC14123 similar to UP|Q93X25 (Q93X25) 2-Cys peroxiredoxin, partial (89%)
Length = 1130
Score = 434 bits (1115), Expect = e-122
Identities = 227/263 (86%), Positives = 234/263 (88%), Gaps = 1/263 (0%)
Frame = +2
Query: 4 CSAPSASLLSSNPNILFSPKLSSPPHLSSLSIPNASNSLPKLRTSLP-LSLNRFTSSRRS 62
CSA S L NP LFSPK ++ SLSIPN+ N L SLP +SL R + SRRS
Sbjct: 65 CSAASLFSLKLNPTPLFSPKPTT-----SLSIPNSLNPLSTKPFSLPSVSLTRPSHSRRS 229
Query: 63 FVVRASSELPLVGNAAPDFEAEAVFDQEFIKVKLSEYIGKKYVILFFYPLDFTFVCPTEI 122
F+VRA+SELPLVGN APDFEAEAVFDQEFIKVKLSEYIGKKYVILFFYPLDFTFVCPTEI
Sbjct: 230 FLVRATSELPLVGNTAPDFEAEAVFDQEFIKVKLSEYIGKKYVILFFYPLDFTFVCPTEI 409
Query: 123 TAFSDRHAEFAELNTEILGVSVDSVFSHLAWVQTDRKSGGLGDLNYPLVSDVTKSISKSY 182
TAFSDRHAEF LNTEILGVSVDSVFSHLAWVQTDRKSGGLGDLNYPLVSDVTKSISKSY
Sbjct: 410 TAFSDRHAEFEALNTEILGVSVDSVFSHLAWVQTDRKSGGLGDLNYPLVSDVTKSISKSY 589
Query: 183 GVLIPDQGIALRGLFIIDKEGVIQHSTINNLGIGRSVDETKRTLQALQYVQENPDEVCPA 242
GVLIPDQGIALRGLFIIDKEGVIQHSTINNL IGRSVDETKRTLQALQYVQENPDEVCPA
Sbjct: 590 GVLIPDQGIALRGLFIIDKEGVIQHSTINNLAIGRSVDETKRTLQALQYVQENPDEVCPA 769
Query: 243 GWKPGEKSMKPDPKLSKEYFSAV 265
GWKPGEKSMKPDPKLSKEYFSAV
Sbjct: 770 GWKPGEKSMKPDPKLSKEYFSAV 838
>TC8321 similar to UP|AAS46230 (AAS46230) Peroxiredoxin Q, partial (75%)
Length = 873
Score = 75.5 bits (184), Expect = 9e-15
Identities = 67/225 (29%), Positives = 108/225 (47%), Gaps = 26/225 (11%)
Frame = +1
Query: 31 SSLSIPNASNSLPKL-----RTSLP--------------LSLNRFTSSRRSFVVRASSEL 71
SSL++PN ++SLP ++LP L L+ +S SF V +SS
Sbjct: 7 SSLTVPNPTHSLPIFLHPPSSSNLPFPFPQSSSNSQFIGLKLSSSSSHSSSFSVPSSSFR 186
Query: 72 PLV------GNAAPDFEAEAVFDQEFIKVKLSEYIGKKYVILFFYPLDFTFVCPTEITAF 125
+ G+ P F + DQ+ V LS++ GK V+++FYP D + C + AF
Sbjct: 187 GSIVAKVTTGSKPPGFTLK---DQDGKTVSLSKFKGKP-VVVYFYPADESPSCTKQACAF 354
Query: 126 SDRHAEFAELNTEILGVSVDSVFSHLAWVQTDRKSGGLGDLNYPLVSDVTKSISKSYGVL 185
D + +F + E++G+S D SH A+ + R L + L+SD + K +GV
Sbjct: 355 RDSYEKFKKAGAEVVGISGDDSSSHKAFAKKYR-------LPFTLLSDEGNKVRKEWGVP 513
Query: 186 IPDQG-IALRGLFIIDKEGVIQHSTINNLGIGRSVDETKRTLQAL 229
G + R +++DK GV+Q N + +DET + LQ+L
Sbjct: 514 GDFFGALPGRETYVLDKSGVVQLVYNNQFQPEKHIDETLKLLQSL 648
>TC8320 similar to UP|AAQ67661 (AAQ67661) Peroxiredoxin Q, partial (31%)
Length = 418
Score = 36.6 bits (83), Expect = 0.005
Identities = 35/125 (28%), Positives = 58/125 (46%), Gaps = 23/125 (18%)
Frame = +3
Query: 31 SSLSIPNASNSLP-----KLRTSLPL------SLNRFTSSRRSFVVRASSELP------- 72
SSL++PN ++SLP ++LP S ++F + S +S +P
Sbjct: 51 SSLTLPNPNHSLPIFIHSPSSSNLPFPFPQSSSNSQFIGLKLSSTHSSSCSVPSSSFKGS 230
Query: 73 -----LVGNAAPDFEAEAVFDQEFIKVKLSEYIGKKYVILFFYPLDFTFVCPTEITAFSD 127
G+ P F + DQ+ V L+++ GK V+++FYP D T C + AF D
Sbjct: 231 IVAKVSKGSKPPAF---TLRDQDGKTVSLTKFKGKP-VVVYFYPADETPGCTKQACAFRD 398
Query: 128 RHAEF 132
+ +F
Sbjct: 399 SYEKF 413
>TC8161 similar to UP|Q8LBV7 (Q8LBV7) Contains similarity to plastid
ribosomal protein L19, partial (54%)
Length = 1101
Score = 32.3 bits (72), Expect = 0.085
Identities = 26/73 (35%), Positives = 33/73 (44%), Gaps = 1/73 (1%)
Frame = +2
Query: 18 ILFSPKLSSPPHLSSLSIPNASNSLPKLRTSLPLSLNRFTSSRRSFVVRASSELPLVG-N 76
+LF+P SP L S + N TS+ N S++ SFVVRA S + N
Sbjct: 104 LLFAPNQCSPTKLGVSSCLGSRNFPLISSTSISWRCNNPLSAKPSFVVRADSNVDAASDN 283
Query: 77 AAPDFEAEAVFDQ 89
A EAE DQ
Sbjct: 284 AGEVPEAEGSVDQ 322
>TC11925 similar to UP|Q9FKX6 (Q9FKX6) Prolyl 4-hydroxylase, alpha
subunit-like protein, partial (43%)
Length = 613
Score = 30.0 bits (66), Expect = 0.42
Identities = 18/44 (40%), Positives = 23/44 (51%)
Frame = +2
Query: 4 CSAPSASLLSSNPNILFSPKLSSPPHLSSLSIPNASNSLPKLRT 47
CS+PS+S S + FS SP H +L PN + S P L T
Sbjct: 110 CSSPSSSSFSFSS---FSVSSLSPMHPLTLVFPNPTTSTPSLAT 232
>TC16620 weakly similar to UP|Q9FN67 (Q9FN67) Gb|AAD56319.1, partial (17%)
Length = 518
Score = 28.9 bits (63), Expect = 0.94
Identities = 21/58 (36%), Positives = 28/58 (48%), Gaps = 6/58 (10%)
Frame = +3
Query: 5 SAPSASLLSSN------PNILFSPKLSSPPHLSSLSIPNASNSLPKLRTSLPLSLNRF 56
S+PS S S PN S LSS P+ + I ++ SLP L ++ S NRF
Sbjct: 264 SSPSFSPFSEQVTSLPRPNPSISKTLSSNPNSTESKISSSIPSLPHLNSNPSTSTNRF 437
>BP064595
Length = 474
Score = 28.5 bits (62), Expect = 1.2
Identities = 22/47 (46%), Positives = 28/47 (58%)
Frame = +2
Query: 11 LLSSNPNILFSPKLSSPPHLSSLSIPNASNSLPKLRTSLPLSLNRFT 57
L SS P++ SP S PP LS LSI + S P L +S PL+L F+
Sbjct: 269 LSSSTPSLTPSPFSSPPPLLSPLSI-SLLTSYP-LASSPPLTLPPFS 403
>BP072692
Length = 422
Score = 28.1 bits (61), Expect = 1.6
Identities = 18/45 (40%), Positives = 24/45 (53%)
Frame = +2
Query: 8 SASLLSSNPNILFSPKLSSPPHLSSLSIPNASNSLPKLRTSLPLS 52
S++ SS+P +P LSSPP SS P+ S S K+ P S
Sbjct: 212 SSATPSSSPTPSAAPPLSSPP--SSTPSPSPSTSSSKVGPDFPFS 340
>TC8850 similar to UP|RK34_SPIOL (P82244) 50S ribosomal protein L34,
chloroplast precursor, partial (49%)
Length = 659
Score = 28.1 bits (61), Expect = 1.6
Identities = 28/68 (41%), Positives = 34/68 (49%), Gaps = 2/68 (2%)
Frame = +2
Query: 8 SASLLSSNPNILFSPKLSSPPHLSSLSIPNASNSLPKLRTSLPLSLNRFTSS--RRSFVV 65
S SL ++ +L LSSP SSLS P S+SL L L L N T + RR VV
Sbjct: 89 SLSLHNTRSPLLRCSFLSSP---SSLSFP--SSSLSGLSLGLDLISNAGTGNGKRRGLVV 253
Query: 66 RASSELPL 73
RA + L
Sbjct: 254 RAGKKFAL 277
>AV410341
Length = 419
Score = 28.1 bits (61), Expect = 1.6
Identities = 26/93 (27%), Positives = 39/93 (40%)
Frame = +3
Query: 8 SASLLSSNPNILFSPKLSSPPHLSSLSIPNASNSLPKLRTSLPLSLNRFTSSRRSFVVRA 67
S+ LLSS P I F+P SLSIP + + T P TS +
Sbjct: 150 SSLLLSSTPRIQFAP--------PSLSIPTLNPPSSRFSTPHPFRFIGLTSK------PS 287
Query: 68 SSELPLVGNAAPDFEAEAVFDQEFIKVKLSEYI 100
++L A A VF++E + L++Y+
Sbjct: 288 INKLVCQAQKASMAAAVEVFEKEELAASLAKYV 386
>TC9137 UP|CAF02297 (CAF02297) Cysteine-rich polycomb-like protein, complete
Length = 3086
Score = 28.1 bits (61), Expect = 1.6
Identities = 17/74 (22%), Positives = 34/74 (44%)
Frame = +1
Query: 27 PPHLSSLSIPNASNSLPKLRTSLPLSLNRFTSSRRSFVVRASSELPLVGNAAPDFEAEAV 86
PP +S + +S+ L ++++ +SLN+ + ++ + PLV N E +A
Sbjct: 1228 PPSCASTTGMRSSDVLQGMQSTSSISLNKVENMKKYVISSNMDRQPLVDNRNEIHETDAS 1407
Query: 87 FDQEFIKVKLSEYI 100
++ L E I
Sbjct: 1408 LAADYFSPSLKEPI 1449
>TC20211 homologue to UP|Q08150 (Q08150) GTP-binding protein 6, partial
(67%)
Length = 623
Score = 28.1 bits (61), Expect = 1.6
Identities = 24/66 (36%), Positives = 32/66 (48%), Gaps = 10/66 (15%)
Frame = +2
Query: 5 SAPSASLLSSNPNILFSPKLSSPPHLSSLSIPNASNSLP----------KLRTSLPLSLN 54
+ P++ +S+P SPK SS SSLS P+A NSLP LR+ PL+
Sbjct: 242 ATPASGSPTSSPG---SPKTSST---SSLSPPSALNSLPAP*MLTPKSSSLRSGTPLAKR 403
Query: 55 RFTSSR 60
SR
Sbjct: 404 GIVQSR 421
>CB827448
Length = 549
Score = 28.1 bits (61), Expect = 1.6
Identities = 26/93 (27%), Positives = 39/93 (40%)
Frame = +3
Query: 8 SASLLSSNPNILFSPKLSSPPHLSSLSIPNASNSLPKLRTSLPLSLNRFTSSRRSFVVRA 67
S+ LLSS P I F+P SLSIP + + T P TS +
Sbjct: 162 SSLLLSSTPRIQFAP--------PSLSIPTLNPPSSRFSTPHPFRFIGLTSK------PS 299
Query: 68 SSELPLVGNAAPDFEAEAVFDQEFIKVKLSEYI 100
++L A A VF++E + L++Y+
Sbjct: 300 INKLVCQAQKASMAAAVEVFEKEELAASLAKYV 398
>AW720237
Length = 459
Score = 28.1 bits (61), Expect = 1.6
Identities = 17/45 (37%), Positives = 26/45 (57%), Gaps = 8/45 (17%)
Frame = -2
Query: 184 VLIPDQGIALRGLFIIDK--------EGVIQHSTINNLGIGRSVD 220
VL DQ +A+R F +++ +G+I+ ST +LGI SVD
Sbjct: 350 VLNSDQRLAIRASFNLERPELDILLDDGIIELSTDKSLGIENSVD 216
>AW719687
Length = 323
Score = 27.7 bits (60), Expect = 2.1
Identities = 22/53 (41%), Positives = 26/53 (48%), Gaps = 5/53 (9%)
Frame = +3
Query: 5 SAPSASLLSSNPNILFSPKLSSPPHLSSL-----SIPNASNSLPKLRTSLPLS 52
SAPSA L SNP+ L S L PP L ++SLP +R LP S
Sbjct: 117 SAPSAVLPPSNPSRLPSAPLHFPPPKKPLLSAVDGSAVCASSLP*MRFVLPCS 275
>AV409768
Length = 405
Score = 27.3 bits (59), Expect = 2.7
Identities = 24/71 (33%), Positives = 30/71 (41%)
Frame = +3
Query: 8 SASLLSSNPNILFSPKLSSPPHLSSLSIPNASNSLPKLRTSLPLSLNRFTSSRRSFVVRA 67
S+S S+ SP L PP L S P A+ SLPK L + SR S
Sbjct: 150 SSSATSTAAASAVSPPLPPPPQLQSS--PPAAASLPKFHLLLTVLPPSSRLSRSSLSPPF 323
Query: 68 SSELPLVGNAA 78
+ LPL +A
Sbjct: 324 PAHLPLTALSA 356
>TC7822 similar to UP|Q42139 (Q42139) H+-transporting ATP synthase CHAIN9 -
like protein (AT4G32260/F10M6_100) , partial (76%)
Length = 907
Score = 27.3 bits (59), Expect = 2.7
Identities = 28/90 (31%), Positives = 45/90 (49%), Gaps = 9/90 (10%)
Frame = +3
Query: 8 SASLLSSNPNILFSPKLSSPPHLSSLSIPNASNSLPKLRTS---LPLSLNRFTSSRRSFV 64
S +L +S N++ + SS P + L I N+ P+L+ S LPL L + +S +
Sbjct: 81 SQNLQASMANMVMA---SSKPLIPPLPISNSLPPTPRLQISLPKLPLKLKHQIPNLKSSL 251
Query: 65 --VRASSELPLVGNAAP----DFEAEAVFD 88
+ A+S L L+ A P + E A+FD
Sbjct: 252 SSLLAASSLSLLAFAPPSLAEEIEKAALFD 341
>BP043430
Length = 493
Score = 26.9 bits (58), Expect = 3.6
Identities = 20/46 (43%), Positives = 26/46 (56%)
Frame = +3
Query: 8 SASLLSSNPNILFSPKLSSPPHLSSLSIPNASNSLPKLRTSLPLSL 53
S+SLLSS+ SS +SLS P++S SLP S PL+L
Sbjct: 282 SSSLLSSS---------SSSSSSTSLSFPSSSLSLPLPSLSSPLAL 392
>AV421748
Length = 376
Score = 26.9 bits (58), Expect = 3.6
Identities = 22/71 (30%), Positives = 31/71 (42%), Gaps = 10/71 (14%)
Frame = +3
Query: 19 LFSPKLSSPPHLSSLSIPNASNSLPKLR----TSLPLSLNRFTSSRRSFVVRA------S 68
L SP HL++ + P+A++SLP T+L L T + S RA +
Sbjct: 69 LVSPSTPRSRHLATTTTPSATSSLPTAALAQLTTLVLPRTATTKKQSSLAPRARSRSTTT 248
Query: 69 SELPLVGNAAP 79
S PL AP
Sbjct: 249 SPTPLPTQTAP 281
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.317 0.134 0.387
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,581,327
Number of Sequences: 28460
Number of extensions: 62116
Number of successful extensions: 525
Number of sequences better than 10.0: 55
Number of HSP's better than 10.0 without gapping: 511
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 517
length of query: 265
length of database: 4,897,600
effective HSP length: 89
effective length of query: 176
effective length of database: 2,364,660
effective search space: 416180160
effective search space used: 416180160
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 54 (25.4 bits)
Medicago: description of AC146630.11


