

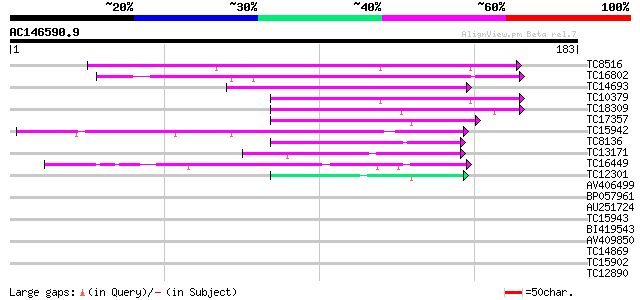
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146590.9 - phase: 0 /pseudo
(183 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC8516 similar to UP|AAP72988 (AAP72988) CDR1, partial (27%) 115 3e-27
TC16802 similar to GB|AAP31963.1|30387595|BT006619 At1g01300 {Ar... 94 1e-20
TC14693 similar to GB|BAB01116.1|11994113|AB026658 CND41, chloro... 74 1e-14
TC10379 weakly similar to UP|Q8H9F4 (Q8H9F4) 41 kDa chloroplast ... 71 1e-13
TC18309 similar to UP|Q9FG60 (Q9FG60) Aspartyl protease-like, pa... 55 9e-09
TC17357 similar to UP|Q9FFC3 (Q9FFC3) Protease-like protein, par... 54 1e-08
TC15942 similar to UP|Q9FL43 (Q9FL43) Nucleoid DNA-binding-like ... 54 2e-08
TC8136 similar to GB|AAB60729.1|2160166|F21M12 F21M12.13 {Arabid... 51 1e-07
TC13171 49 7e-07
TC16449 similar to UP|Q940R4 (Q940R4) AT4g16560/dl4305c, partial... 44 2e-05
TC12301 weakly similar to UP|CAR5_RHINI (P43232) Rhizopuspepsin ... 40 3e-04
AV406499 36 0.003
BP057961 35 0.006
AU251724 35 0.008
TC15943 similar to UP|Q8GT67 (Q8GT67) Xyloglucan-specific fungal... 33 0.038
BI419543 33 0.038
AV409850 33 0.038
TC14869 32 0.065
TC15902 similar to UP|Q948P0 (Q948P0) Aspartic proteinase alpha,... 30 0.32
TC12890 homologue to UP|Q40359 (Q40359) Alfin-1, partial (23%) 29 0.42
>TC8516 similar to UP|AAP72988 (AAP72988) CDR1, partial (27%)
Length = 1603
Score = 115 bits (289), Expect = 3e-27
Identities = 60/150 (40%), Positives = 85/150 (56%), Gaps = 10/150 (6%)
Frame = +1
Query: 26 NGFSVELIHRDSSKSPLYQPTQNKYQHIVNAARRSINRAN-------HFYKTALTNTPQS 78
+ F+++LIH DS SP Y + + Q I NAA RSI+RAN H ++P+
Sbjct: 145 SSFTIDLIHHDSPLSPFYNSSMTRSQLIRNAAMRSISRANQLSLSLSHSLNQLKESSPEP 324
Query: 79 TVIPDHGEYLMTYSVGTPPFKLYGIADTGSDIVWLQCEPC--KECYNQTTPKFKPSKSST 136
+IP++G YLM +GTP + IADTGSD+ W+QC PC +C+ Q TP + P SST
Sbjct: 325 IIIPNNGNYLMRIYIGTPSVERLAIADTGSDLTWVQCSPCDNTKCFAQNTPLYDPLNSST 504
Query: 137 YKNIPCSSDLC-KSGQQGAVCTDQNFCEYS 165
+ +PC S C + VC+D C Y+
Sbjct: 505 FTLLPCDSQPCTQLPYSQYVCSDYGDCIYA 594
>TC16802 similar to GB|AAP31963.1|30387595|BT006619 At1g01300 {Arabidopsis
thaliana;}, partial (28%)
Length = 812
Score = 94.0 bits (232), Expect = 1e-20
Identities = 56/153 (36%), Positives = 75/153 (48%), Gaps = 15/153 (9%)
Frame = +3
Query: 29 SVELIHRDSSKSPLYQPTQNKYQHIVNAARRSINRANHFYKT---ALTNTPQ-------- 77
S++L H D+ S T+ Q +R RA F+ T AL N+
Sbjct: 246 SLQLHHIDALSS-----TKTPEQLFHLRLQRDAARAESFFTTSAAALNNSHAPRSGAGFS 410
Query: 78 ----STVIPDHGEYLMTYSVGTPPFKLYGIADTGSDIVWLQCEPCKECYNQTTPKFKPSK 133
S + GEY VGTP +Y + DTGSD+VWLQC PC++CY+Q P F PSK
Sbjct: 411 SSIISGLAQGSGEYFTRIGVGTPARYVYMVLDTGSDVVWLQCAPCRKCYSQADPVFDPSK 590
Query: 134 SSTYKNIPCSSDLCKSGQQGAVCTDQNFCEYSI 166
S TY IPC + LC+ A C + C+Y +
Sbjct: 591 SRTYAGIPCGAPLCRR-LDSAGCNSKKVCQYQV 686
>TC14693 similar to GB|BAB01116.1|11994113|AB026658 CND41, chloroplast
nucleoid DNA binding protein-like {Arabidopsis
thaliana;} , partial (58%)
Length = 2067
Score = 73.9 bits (180), Expect = 1e-14
Identities = 33/79 (41%), Positives = 44/79 (54%)
Frame = +1
Query: 71 ALTNTPQSTVIPDHGEYLMTYSVGTPPFKLYGIADTGSDIVWLQCEPCKECYNQTTPKFK 130
AL+ S GEY VG P Y + DTGSD+ WLQC+PC +CY Q+ P F
Sbjct: 463 ALSTPVSSGTGQGSGEYFSRVGVGQPAKPFYMVLDTGSDVNWLQCKPCADCYQQSDPIFD 642
Query: 131 PSKSSTYKNIPCSSDLCKS 149
P+ SSTY + C + C++
Sbjct: 643 PTTSSTYGPLTCDAQQCQA 699
>TC10379 weakly similar to UP|Q8H9F4 (Q8H9F4) 41 kDa chloroplast nucleoid
DNA binding protein (CND41), partial (43%)
Length = 1002
Score = 70.9 bits (172), Expect = 1e-13
Identities = 35/88 (39%), Positives = 46/88 (51%), Gaps = 6/88 (6%)
Frame = +3
Query: 85 GEYLMTYSVGTPPFKLYGIADTGSDIVWLQCEPC-KECYNQTTPKFKPSKSSTYKNIPCS 143
G Y + +GTP L + DTGSD+ W QC+PC + CY Q P F P +S++Y NI C+
Sbjct: 423 GNYFVVVGLGTPKSDLSLVFDTGSDLTWTQCQPCARSCYKQQDPIFDPKRSTSYYNITCT 602
Query: 144 SDLC-----KSGQQGAVCTDQNFCEYSI 166
S C +G A T C Y I
Sbjct: 603 SSDCTQLTSATGNDPACATITKACVYGI 686
>TC18309 similar to UP|Q9FG60 (Q9FG60) Aspartyl protease-like, partial (14%)
Length = 624
Score = 54.7 bits (130), Expect = 9e-09
Identities = 28/89 (31%), Positives = 42/89 (46%), Gaps = 7/89 (7%)
Frame = +2
Query: 85 GEYLMTYSVGTPPFKLYGIADTGSDIVWLQCEPCKECYNQT-----TPKFKPSKSSTYKN 139
G Y +G+P Y DTGSDI+W+ C C C ++ + P +S T +
Sbjct: 248 GLYFTKIGLGSPSKDYYVQVDTGSDILWVNCVECTRCPRKSDIGIGLTLYDPKRSKTSEF 427
Query: 140 IPCSSDLCKSGQQGAV--CTDQNFCEYSI 166
+ C + C S +G + C +N C YSI
Sbjct: 428 VSCEHNFCSSTYEGRILGCKAENPCPYSI 514
>TC17357 similar to UP|Q9FFC3 (Q9FFC3) Protease-like protein, partial (24%)
Length = 577
Score = 54.3 bits (129), Expect = 1e-08
Identities = 27/73 (36%), Positives = 34/73 (45%), Gaps = 5/73 (6%)
Frame = +1
Query: 85 GEYLMTYSVGTPPFKLYGIADTGSDIVWLQCEPCKECYNQTTPK-----FKPSKSSTYKN 139
G Y +GTPP + Y DTGSD++W+ C C C + + F P SST
Sbjct: 313 GLYFTRVQIGTPPVEFYVQIDTGSDVLWVSCSSCNGCPQTSGLQIQLDFFDPGHSSTSSL 492
Query: 140 IPCSSDLCKSGQQ 152
I CS C G Q
Sbjct: 493 ISCSDQRCNGGIQ 531
>TC15942 similar to UP|Q9FL43 (Q9FL43) Nucleoid DNA-binding-like protein,
partial (66%)
Length = 973
Score = 53.5 bits (127), Expect = 2e-08
Identities = 40/155 (25%), Positives = 68/155 (43%), Gaps = 9/155 (5%)
Frame = +1
Query: 3 TCSLLILFYFSLCFIISL----SHALNNGFSVELIHRDSSKSPLYQPTQNKYQH-IVNAA 57
T SLL LF SL ++ SH ++G ++++ H S SP P ++ ++
Sbjct: 10 TLSLLFLFLSSLVQGLTYPKCDSH--DHGSTLQVFHVFSPCSPFRPPKPLSWEESVLQMQ 183
Query: 58 RRSINRANHFYKT----ALTNTPQSTVIPDHGEYLMTYSVGTPPFKLYGIADTGSDIVWL 113
+ I R ++ I Y++ +G PP L DT +D W+
Sbjct: 184 AKDITRVQFLVSLVAGRSIVPIASGRQIIQSPTYIVRAKIGNPPQTLLMAMDTSNDAAWV 363
Query: 114 QCEPCKECYNQTTPKFKPSKSSTYKNIPCSSDLCK 148
C C C ++ F P KS+T+K++ C++ CK
Sbjct: 364 PCSGCDGC---SSALFAPEKSTTFKSVGCAAAECK 459
>TC8136 similar to GB|AAB60729.1|2160166|F21M12 F21M12.13 {Arabidopsis
thaliana;}, partial (70%)
Length = 1659
Score = 50.8 bits (120), Expect = 1e-07
Identities = 23/63 (36%), Positives = 33/63 (51%)
Frame = +1
Query: 85 GEYLMTYSVGTPPFKLYGIADTGSDIVWLQCEPCKECYNQTTPKFKPSKSSTYKNIPCSS 144
G Y++ +GTP L+ + DT +D ++ C C C T P F P S+TY + CS
Sbjct: 352 GNYIVRVKIGTPGQLLFMVLDTSTDEAFVPCSGCTGCSTTTAP-FSPKASTTYSPLDCSV 528
Query: 145 DLC 147
LC
Sbjct: 529 PLC 537
>TC13171
Length = 598
Score = 48.5 bits (114), Expect = 7e-07
Identities = 26/73 (35%), Positives = 42/73 (56%), Gaps = 1/73 (1%)
Frame = +3
Query: 76 PQSTVIPDHGEYL-MTYSVGTPPFKLYGIADTGSDIVWLQCEPCKECYNQTTPKFKPSKS 134
PQ+ + H L +T +VG+PP + + DTGS++ WL C+ K + ++ F P S
Sbjct: 273 PQNKLSFHHNVSLTVTLTVGSPPQNVTMVLDTGSELSWLHCK--KAPNSNSSSIFNPPLS 446
Query: 135 STYKNIPCSSDLC 147
S+Y PC+S +C
Sbjct: 447 SSYNPTPCTSPVC 485
>TC16449 similar to UP|Q940R4 (Q940R4) AT4g16560/dl4305c, partial (18%)
Length = 613
Score = 43.5 bits (101), Expect = 2e-05
Identities = 41/142 (28%), Positives = 59/142 (40%), Gaps = 4/142 (2%)
Frame = +3
Query: 12 FSLCFIISLSHALNNGFSVELIHRDSSKSPLYQPTQNKYQHIVNA-ARRSINRANHFYKT 70
F LCF++ +SH+ + L H SKS N H++ + RS R H +
Sbjct: 87 FLLCFMLCISHSYQMVL-LPLTH-SISKSQF-----NSTHHLLKTTSTRSAARFQHHHHR 245
Query: 71 ALTNTPQSTVIPDHGEYLMTYSVGTPPFKLYGIADTGSDIVWLQCEP--CKECYNQ-TTP 127
+ + P L G+ P LY DTGSD+VW C P C C + TP
Sbjct: 246 HQQHQLSLPLSPGSDYTLSLNFGGSQPITLY--MDTGSDLVWFPCAPFECMLCEGKPDTP 419
Query: 128 KFKPSKSSTYKNIPCSSDLCKS 149
KP+ + + C S C +
Sbjct: 420 --KPANITKAHTVSCQSPACSA 479
>TC12301 weakly similar to UP|CAR5_RHINI (P43232) Rhizopuspepsin 5 precursor
(Aspartate protease) , partial (8%)
Length = 501
Score = 39.7 bits (91), Expect = 3e-04
Identities = 27/94 (28%), Positives = 36/94 (37%), Gaps = 30/94 (31%)
Frame = +2
Query: 85 GEYLMTYSVGTPPFKLYGIADTGSDIVWLQCEPCKECYNQTTPK---------------- 128
GEY + VGTP K + ADTGS+ W + +N+T K
Sbjct: 14 GEYFVQVKVGTPGQKFWLAADTGSEFTWF--NSVHKTHNKTQTKNHGGGGHKKHKGKLRK 187
Query: 129 --------------FKPSKSSTYKNIPCSSDLCK 148
F P +S T+K + CSS CK
Sbjct: 188 RGKKVRHNNPCNGVFCPQRSRTFKTVTCSSRKCK 289
>AV406499
Length = 400
Score = 36.2 bits (82), Expect = 0.003
Identities = 40/143 (27%), Positives = 60/143 (40%), Gaps = 15/143 (10%)
Frame = +1
Query: 7 LILFYFSLCFIISL-----SHALNNGFSVELIHRDSSKSPLYQPTQNK--YQHIVNAARR 59
L LF FS F++S+ S+ NN + L +S PL T +K Y + ++++
Sbjct: 4 LFLFLFSF-FLLSVNPSVQSNTTNNPKPLSLSFPLASL-PLSTNTSSKLLYTSLFSSSKH 177
Query: 60 SINRANHFYKTALTNTPQSTVIPDHGEYLMTYS--------VGTPPFKLYGIADTGSDIV 111
+ HF TP S+ + + YS +GTPP + DTGS +
Sbjct: 178 T---TPHF------KTPPSSSSQHSDRFSLKYSMALIVSLPIGTPPQVQQMVLDTGSQLS 330
Query: 112 WLQCEPCKECYNQTTPKFKPSKS 134
W+QC T F PS S
Sbjct: 331 WIQCRHHNRNAPPPTASFNPSLS 399
>BP057961
Length = 398
Score = 35.4 bits (80), Expect = 0.006
Identities = 16/30 (53%), Positives = 17/30 (56%)
Frame = +3
Query: 92 SVGTPPFKLYGIADTGSDIVWLQCEPCKEC 121
SVGTPP DTGSD+ WL C C C
Sbjct: 234 SVGTPPLWFVVALDTGSDLFWLPCN-CTSC 320
>AU251724
Length = 399
Score = 35.0 bits (79), Expect = 0.008
Identities = 23/65 (35%), Positives = 35/65 (53%), Gaps = 2/65 (3%)
Frame = +1
Query: 57 ARRSINRANHFYKTALTNTPQSTVIPDHGEYLMTYSVGTPPFKLYGI-ADTGSDIVWLQC 115
A+ N N +A+ Q V P G Y + ++G PP K Y + D+GSD+ W++C
Sbjct: 211 AKNPRNTENRLGSSAVFKV-QGNVYP-LGYYTVFINIGHPP-KFYDLDIDSGSDLTWIEC 381
Query: 116 E-PCK 119
+ PCK
Sbjct: 382 DGPCK 396
>TC15943 similar to UP|Q8GT67 (Q8GT67) Xyloglucan-specific fungal
endoglucanase inhibitor protein precursor, partial (15%)
Length = 968
Score = 32.7 bits (73), Expect = 0.038
Identities = 22/71 (30%), Positives = 29/71 (39%)
Frame = +2
Query: 87 YLMTYSVGTPPFKLYGIADTGSDIVWLQCEPCKECYNQTTPKFKPSKSSTYKNIPCSSDL 146
+ + VGTP + D G I+W C YN SSTY+ + C S
Sbjct: 194 FYTSVGVGTPRHNFDLVIDLGGPILWYD---CNNHYN----------SSTYRPVSCDSKS 334
Query: 147 CKSGQQGAVCT 157
C + GA CT
Sbjct: 335 CSN--TGAGCT 361
>BI419543
Length = 585
Score = 32.7 bits (73), Expect = 0.038
Identities = 19/54 (35%), Positives = 25/54 (46%), Gaps = 3/54 (5%)
Frame = +2
Query: 86 EYLMTYSVGTPPFKLYGIADTGSDIVWLQCEPCK---ECYNQTTPKFKPSKSST 136
+Y ++GTPP I DTGS +W+ C CY K K SK+ T
Sbjct: 389 QYYGEIAIGTPPQTFDVIFDTGSSNLWVPSSKCYFSIACYTHHWYKSKKSKTYT 550
>AV409850
Length = 419
Score = 32.7 bits (73), Expect = 0.038
Identities = 13/29 (44%), Positives = 18/29 (61%)
Frame = +1
Query: 85 GEYLMTYSVGTPPFKLYGIADTGSDIVWL 113
G Y +GTPP + Y DTGSD++W+
Sbjct: 331 GLYYTKVKLGTPPREFYVQIDTGSDVLWV 417
>TC14869
Length = 828
Score = 32.0 bits (71), Expect = 0.065
Identities = 25/84 (29%), Positives = 39/84 (45%), Gaps = 5/84 (5%)
Frame = +3
Query: 73 TNTPQSTVIP---DHG--EYLMTYSVGTPPFKLYGIADTGSDIVWLQCEPCKECYNQTTP 127
+++P S ++P DH +YL T S GTP + D G + W +C ++ TP
Sbjct: 219 SSSPISLLLPITKDHSTLQYLTTLSTGTPSAPTNLVLDLGGSLPWF------DCASRHTP 380
Query: 128 KFKPSKSSTYKNIPCSSDLCKSGQ 151
SS+ + IP S C + Q
Sbjct: 381 ------SSSLRTIPHRSLQCLTHQ 434
>TC15902 similar to UP|Q948P0 (Q948P0) Aspartic proteinase alpha, partial
(25%)
Length = 517
Score = 29.6 bits (65), Expect = 0.32
Identities = 14/37 (37%), Positives = 19/37 (50%)
Frame = +3
Query: 86 EYLMTYSVGTPPFKLYGIADTGSDIVWLQCEPCKECY 122
+Y +GTPP I DTGS +W+ P +CY
Sbjct: 387 QYFGEIGIGTPPQTFTVIFDTGSSNLWV---PSSKCY 488
>TC12890 homologue to UP|Q40359 (Q40359) Alfin-1, partial (23%)
Length = 460
Score = 29.3 bits (64), Expect = 0.42
Identities = 11/38 (28%), Positives = 19/38 (49%)
Frame = +3
Query: 107 GSDIVWLQCEPCKECYNQTTPKFKPSKSSTYKNIPCSS 144
G+D W+ C+ C+ ++ K P+K+ K C S
Sbjct: 78 GTDEFWICCDMCERWFHGKCVKITPAKAEHIKQYKCPS 191
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.322 0.136 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,081,692
Number of Sequences: 28460
Number of extensions: 65655
Number of successful extensions: 454
Number of sequences better than 10.0: 56
Number of HSP's better than 10.0 without gapping: 445
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 448
length of query: 183
length of database: 4,897,600
effective HSP length: 85
effective length of query: 98
effective length of database: 2,478,500
effective search space: 242893000
effective search space used: 242893000
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 52 (24.6 bits)
Medicago: description of AC146590.9


