

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146590.13 + phase: 0
(314 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
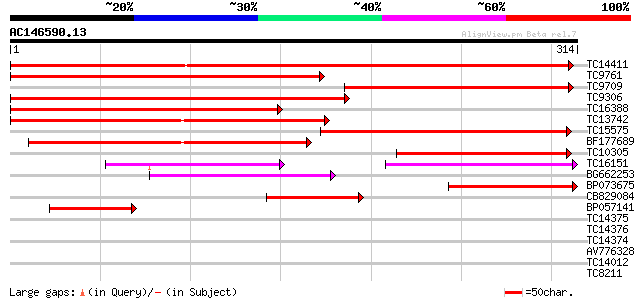
Score E
Sequences producing significant alignments: (bits) Value
TC14411 similar to UP|O65848 (O65848) Annexin, complete 442 e-125
TC9761 similar to UP|O65848 (O65848) Annexin, partial (55%) 240 2e-64
TC9709 similar to PIR|S56674|S56674 annexin homolog RJ4 (clone R... 219 4e-58
TC9306 similar to UP|Q42922 (Q42922) Annexin (Fragment), partial... 173 3e-44
TC16388 similar to UP|O82090 (O82090) Fiber annexin, partial (48%) 140 3e-34
TC13742 similar to UP|Q9ZR53 (Q9ZR53) Annexin-like protein, part... 132 1e-31
TC15575 similar to UP|O82090 (O82090) Fiber annexin, partial (45%) 123 4e-29
BF177689 107 3e-24
TC10305 homologue to UP|Q42922 (Q42922) Annexin (Fragment), part... 91 3e-19
TC16151 similar to UP|Q9ZR53 (Q9ZR53) Annexin-like protein, part... 82 2e-16
BG662253 81 2e-16
BP073675 68 2e-12
CB829084 60 4e-10
BP057141 60 5e-10
TC14375 similar to UP|O65848 (O65848) Annexin, partial (16%) 35 0.002
TC14376 similar to UP|O65848 (O65848) Annexin, partial (14%) 34 0.005
TC14374 similar to UP|O65848 (O65848) Annexin, partial (5%) 35 0.022
AV776328 33 0.082
TC14012 similar to UP|Q8GSM9 (Q8GSM9) Squalene monooxygenase 2 ... 27 3.4
TC8211 homologue to UP|PDX1_PHAVU (Q9FT25) Probable pyridoxin bi... 26 7.7
>TC14411 similar to UP|O65848 (O65848) Annexin, complete
Length = 1306
Score = 442 bits (1137), Expect = e-125
Identities = 227/312 (72%), Positives = 265/312 (84%)
Frame = +3
Query: 1 MATLIAPMNHSPKEDADVLWKAVKGWGTDESAIIAIMGQRNAVQRQQIRQAYQDIYQEDL 60
MATLIAP NHSP+ DA+ L KA +GWGTDE+ +I+I+G RN QRQ IR+AY++IYQEDL
Sbjct: 48 MATLIAPSNHSPQADAESLKKAFEGWGTDENLVISILGHRNVHQRQAIRRAYEEIYQEDL 227
Query: 61 IKRLESELSGNFEKAMYRWILDPADRYAVLANVAIKSINKDYHVIVEIASVLQPQELLAV 120
+KRLESE+ G+ EKA+YRW L+ ADR AVL NV IKS K+YHVIVEI+SVL P+ELLAV
Sbjct: 228 VKRLESEIKGDLEKAVYRWNLEHADRDAVLINVVIKS-GKNYHVIVEISSVLSPEELLAV 404
Query: 121 RHAYHNRYKNSLEEDVAAHTSGYHRQLLVGLVSSFRYDGVEINPILAKHEADILHEAVKN 180
R AY NRYK+SLEEDVAAHTSG+ RQLLVGLV+SFRY G EIN LA+ EA+ILHEAVK
Sbjct: 405 RRAYLNRYKHSLEEDVAAHTSGHLRQLLVGLVTSFRYVGEEINAKLAQSEAEILHEAVKE 584
Query: 181 KKGNIEEVIRILITRSKTQLKATFNRYRDDHGFSISKKLLNEASDDFLKAVHVAIRCIDD 240
KKG+ EE IRIL TRSKTQL ATFNRYR+ HG SI+KKLL+E SDDF K ++ AIRC +D
Sbjct: 585 KKGSHEEAIRILTTRSKTQLIATFNRYREIHGTSITKKLLDEKSDDFQKGLYTAIRCFND 764
Query: 241 HKKYYEKVLRGALKRIGTDEDGLTRVVITRAEKDLKDIKELYYKRNSVHLEDTVAKEISG 300
H KYYEKV+R A+K+ GTDED LTRV+++RAEKDLK I ++YYKRNSVHLED VAKEISG
Sbjct: 765 HIKYYEKVVRDAIKKSGTDEDALTRVIVSRAEKDLKLISDVYYKRNSVHLEDAVAKEISG 944
Query: 301 DYKKFLLTLLGK 312
DYKKFLLTLLGK
Sbjct: 945 DYKKFLLTLLGK 980
>TC9761 similar to UP|O65848 (O65848) Annexin, partial (55%)
Length = 558
Score = 240 bits (613), Expect = 2e-64
Identities = 124/174 (71%), Positives = 143/174 (81%)
Frame = +1
Query: 1 MATLIAPMNHSPKEDADVLWKAVKGWGTDESAIIAIMGQRNAVQRQQIRQAYQDIYQEDL 60
MATLIA HSP DA+ L A +GWGTDE +IAI+G R+ QRQQIR+AY+++YQED+
Sbjct: 37 MATLIATTQHSPVADAEALRNAFQGWGTDEKTVIAILGHRSVHQRQQIRKAYEELYQEDI 216
Query: 61 IKRLESELSGNFEKAMYRWILDPADRYAVLANVAIKSINKDYHVIVEIASVLQPQELLAV 120
IKRLESELSG+ EKA+YRW+L+P DR AVLANVAIKS K Y+VIVEIA+VL P+E+LAV
Sbjct: 217 IKRLESELSGDIEKAVYRWMLEPTDRDAVLANVAIKSGGKGYNVIVEIATVLSPEEVLAV 396
Query: 121 RHAYHNRYKNSLEEDVAAHTSGYHRQLLVGLVSSFRYDGVEINPILAKHEADIL 174
R AYHNRYK SLEEDVAAHT+G RQL VGLVSSFRY G EIN LAK EADIL
Sbjct: 397 RRAYHNRYKRSLEEDVAAHTTGDLRQLWVGLVSSFRYGGDEINARLAKTEADIL 558
>TC9709 similar to PIR|S56674|S56674 annexin homolog RJ4 (clone RJ4) -
garden strawberry (fragment) {Fragaria x
ananassa;}, partial (48%)
Length = 577
Score = 219 bits (559), Expect = 4e-58
Identities = 107/127 (84%), Positives = 118/127 (92%)
Frame = +2
Query: 186 EEVIRILITRSKTQLKATFNRYRDDHGFSISKKLLNEASDDFLKAVHVAIRCIDDHKKYY 245
EE IRIL TRSKTQL ATFNRYRDDHG SI+KKLL+ ASDDF KA+H AIRCI+DH+KYY
Sbjct: 5 EEAIRILTTRSKTQLVATFNRYRDDHGISITKKLLDNASDDFHKALHTAIRCINDHQKYY 184
Query: 246 EKVLRGALKRIGTDEDGLTRVVITRAEKDLKDIKELYYKRNSVHLEDTVAKEISGDYKKF 305
EKVLR ALKR+G+DEDGLTRVV+TRAEKDLK+IKELYYKRNSVHLED VAKE+SGDYKKF
Sbjct: 185 EKVLRNALKRVGSDEDGLTRVVVTRAEKDLKEIKELYYKRNSVHLEDAVAKELSGDYKKF 364
Query: 306 LLTLLGK 312
+LTLLGK
Sbjct: 365 ILTLLGK 385
>TC9306 similar to UP|Q42922 (Q42922) Annexin (Fragment), partial (59%)
Length = 662
Score = 173 bits (439), Expect = 3e-44
Identities = 93/189 (49%), Positives = 123/189 (64%), Gaps = 1/189 (0%)
Frame = +1
Query: 1 MATLIAPMN-HSPKEDADVLWKAVKGWGTDESAIIAIMGQRNAVQRQQIRQAYQDIYQED 59
MATL P SP ED++ L KA +GWGT+E II+I+ RNA QR+ I + Y Y ED
Sbjct: 94 MATLKVPSQVPSPAEDSEQLRKAFQGWGTNEDLIISILAHRNAAQRKLIHETYSQTYGED 273
Query: 60 LIKRLESELSGNFEKAMYRWILDPADRYAVLANVAIKSINKDYHVIVEIASVLQPQELLA 119
L+ L+ ELS +FE+A+ W L PA+R A L N A K + K+ +++EIAS +L
Sbjct: 274 LLTDLDKELSSDFERAVVLWTLGPAERDAFLVNEATKRLTKNNWILMEIASTRSSLDLFK 453
Query: 120 VRHAYHNRYKNSLEEDVAAHTSGYHRQLLVGLVSSFRYDGVEINPILAKHEADILHEAVK 179
+ AY R+K S+EEDVA HTSG R+LLV LV +FRYDG E+N ILAK EA +LHE +
Sbjct: 454 AKQAYQARFKRSIEEDVAYHTSGDIRKLLVPLVGTFRYDGDEVNMILAKSEAKLLHEKIA 633
Query: 180 NKKGNIEEV 188
K N E++
Sbjct: 634 EKAYNHEDL 660
>TC16388 similar to UP|O82090 (O82090) Fiber annexin, partial (48%)
Length = 574
Score = 140 bits (353), Expect = 3e-34
Identities = 75/152 (49%), Positives = 98/152 (64%), Gaps = 1/152 (0%)
Frame = +1
Query: 1 MATLIAPMNHSP-KEDADVLWKAVKGWGTDESAIIAIMGQRNAVQRQQIRQAYQDIYQED 59
MATL P P +D + L KA GWGT+E II+I+G RNA QR+ IR+ Y + Y ED
Sbjct: 118 MATLRVPQTPPPVADDCEQLRKAFSGWGTNEDLIISILGHRNAAQRKLIRETYAETYGED 297
Query: 60 LIKRLESELSGNFEKAMYRWILDPADRYAVLANVAIKSINKDYHVIVEIASVLQPQELLA 119
L+K L+ EL+ +FE+ ++ W LD A+R A LAN A K V+VEIA +++ A
Sbjct: 298 LLKALDKELTSDFERLVHLWALDSAERDAFLANEATKRWTSSNQVLVEIACTRSSEQMFA 477
Query: 120 VRHAYHNRYKNSLEEDVAAHTSGYHRQLLVGL 151
VR AYH YK SLEEDVA HT+G R+LL+ L
Sbjct: 478 VRKAYHALYKKSLEEDVAHHTTGDFRKLLLPL 573
>TC13742 similar to UP|Q9ZR53 (Q9ZR53) Annexin-like protein, partial (53%)
Length = 612
Score = 132 bits (331), Expect = 1e-31
Identities = 68/178 (38%), Positives = 114/178 (63%), Gaps = 1/178 (0%)
Frame = +2
Query: 1 MATL-IAPMNHSPKEDADVLWKAVKGWGTDESAIIAIMGQRNAVQRQQIRQAYQDIYQED 59
M+TL + P+ SP++DA L+ A KG+G D S +I I+ R+A QR I+Q Y+ +Y D
Sbjct: 77 MSTLNVPPIPPSPRDDAIQLYSAFKGFGCDTSIVINILAHRDATQRAYIQQEYRTMYSGD 256
Query: 60 LIKRLESELSGNFEKAMYRWILDPADRYAVLANVAIKSINKDYHVIVEIASVLQPQELLA 119
L+KRL SELSG E A+ W+ DPA R A++ ++ ++ K+ E+ P +L
Sbjct: 257 LLKRLSSELSGKLETAVLLWMHDPAGRDAIILKQSL-TVAKNLETATEVICSRTPSQLQY 433
Query: 120 VRHAYHNRYKNSLEEDVAAHTSGYHRQLLVGLVSSFRYDGVEINPILAKHEADILHEA 177
+R YH+++ LE D+ +TSG H+++L+ VS+ R++G E+N +A+ +A++L++A
Sbjct: 434 LRQIYHSKFGTYLEHDIERNTSGDHKKILLAYVSTPRHEGPEVNREMAQKDANVLYKA 607
>TC15575 similar to UP|O82090 (O82090) Fiber annexin, partial (45%)
Length = 747
Score = 123 bits (309), Expect = 4e-29
Identities = 61/139 (43%), Positives = 91/139 (64%)
Frame = +1
Query: 173 ILHEAVKNKKGNIEEVIRILITRSKTQLKATFNRYRDDHGFSISKKLLNEASDDFLKAVH 232
+LHE + +K N +++IRIL TRS+ Q+ AT N Y+D G I+K L E D++L +
Sbjct: 4 LLHEKITDKAYNDDDLIRILATRSRAQINATLNHYKDAFGKDINKDLKAEPKDEYLSLLR 183
Query: 233 VAIRCIDDHKKYYEKVLRGALKRIGTDEDGLTRVVITRAEKDLKDIKELYYKRNSVHLED 292
++C+ +KY+EK +R ++ + GTDE LTRVV RAE DLK I Y +R+S+ L+
Sbjct: 184 ATVKCLVRPEKYFEKFIRLSINKRGTDEGALTRVVAPRAEIDLKIIANEYQRRSSIPLDR 363
Query: 293 TVAKEISGDYKKFLLTLLG 311
+ K+ +GDY+K LL LLG
Sbjct: 364 AIIKDTNGDYEKMLLALLG 420
>BF177689
Length = 509
Score = 107 bits (266), Expect = 3e-24
Identities = 57/157 (36%), Positives = 96/157 (60%)
Frame = +3
Query: 11 SPKEDADVLWKAVKGWGTDESAIIAIMGQRNAVQRQQIRQAYQDIYQEDLIKRLESELSG 70
+P++DA L++A KG+G D +A+I I+ R+A QR ++Q Y+ Y E+L KRL SELSG
Sbjct: 24 TPRDDAMQLYRAFKGFGCDTTAVINILAHRDATQRAYLQQEYKATYAEELSKRLISELSG 203
Query: 71 NFEKAMYRWILDPADRYAVLANVAIKSINKDYHVIVEIASVLQPQELLAVRHAYHNRYKN 130
E A+ W+ DPA R A + + +++ ++ E+ P +L ++ YH+++
Sbjct: 204 KLETAVLLWMHDPAGRDATIIRQCL-AVDMNFEGATEVICSRTPSQLQYLKQIYHSKFGV 380
Query: 131 SLEEDVAAHTSGYHRQLLVGLVSSFRYDGVEINPILA 167
LE D+ A TSG +++L+ VS+ R +G E+N +A
Sbjct: 381 YLEHDIEATTSGDLKKILLAYVSTPRPEGPEVNREIA 491
>TC10305 homologue to UP|Q42922 (Q42922) Annexin (Fragment), partial (32%)
Length = 498
Score = 90.5 bits (223), Expect = 3e-19
Identities = 46/97 (47%), Positives = 65/97 (66%)
Frame = +3
Query: 215 ISKKLLNEASDDFLKAVHVAIRCIDDHKKYYEKVLRGALKRIGTDEDGLTRVVITRAEKD 274
I K L ++ D++L + I+ + +KY+E++LR A+ + GTDE LTRVV TRAE D
Sbjct: 6 IDKDLETDSDDEYLNLLRATIKSLTYPEKYFEELLRLAINKTGTDEWALTRVVTTRAEVD 185
Query: 275 LKDIKELYYKRNSVHLEDTVAKEISGDYKKFLLTLLG 311
L+ I E Y KRNSV L+ +A + SGDY+K LL L+G
Sbjct: 186 LQKIAEEYQKRNSVPLDRAIANDTSGDYQKILLALMG 296
>TC16151 similar to UP|Q9ZR53 (Q9ZR53) Annexin-like protein, partial (31%)
Length = 587
Score = 81.6 bits (200), Expect = 2e-16
Identities = 43/106 (40%), Positives = 61/106 (56%)
Frame = +2
Query: 209 DDHGFSISKKLLNEASDDFLKAVHVAIRCIDDHKKYYEKVLRGALKRIGTDEDGLTRVVI 268
D +G ++K + NE S F A+ +C + KY+ KVL A+K +GT++ L RV++
Sbjct: 5 DMYGHKLNKAIKNETSGIFAHALLTIFQCAVNPAKYFAKVLHKAMKGLGTNDTTLIRVIV 184
Query: 269 TRAEKDLKDIKELYYKRNSVHLEDTVAKEISGDYKKFLLTLLGKGH 314
TR E D++ IK Y K+ L D V E SG Y+ FLL LLG H
Sbjct: 185 TRTEIDMQYIKAEYLKKYKKTLNDAVHSETSGHYRAFLLALLGPNH 322
Score = 46.2 bits (108), Expect = 7e-06
Identities = 25/102 (24%), Positives = 55/102 (53%), Gaps = 3/102 (2%)
Frame = +2
Query: 54 DIYQEDLIKRLESELSGNFEKAM---YRWILDPADRYAVLANVAIKSINKDYHVIVEIAS 110
D+Y L K +++E SG F A+ ++ ++PA +A + + A+K + + ++ +
Sbjct: 5 DMYGHKLNKAIKNETSGIFAHALLTIFQCAVNPAKYFAKVLHKAMKGLGTNDTTLIRVIV 184
Query: 111 VLQPQELLAVRHAYHNRYKNSLEEDVAAHTSGYHRQLLVGLV 152
++ ++ Y +YK +L + V + TSG++R L+ L+
Sbjct: 185 TRTEIDMQYIKAEYLKKYKKTLNDAVHSETSGHYRAFLLALL 310
Score = 37.7 bits (86), Expect = 0.003
Identities = 27/99 (27%), Positives = 48/99 (48%)
Frame = +2
Query: 125 HNRYKNSLEEDVAAHTSGYHRQLLVGLVSSFRYDGVEINPILAKHEADILHEAVKNKKGN 184
H+ Y + L + + TSG L+ + +NP AK+ A +LH+A+K N
Sbjct: 2 HDMYGHKLNKAIKNETSGIFAHALLTIFQC------AVNP--AKYFAKVLHKAMKGLGTN 157
Query: 185 IEEVIRILITRSKTQLKATFNRYRDDHGFSISKKLLNEA 223
+IR+++TR++ ++ Y + KK LN+A
Sbjct: 158 DTTLIRVIVTRTEIDMQYIKAEYLKKY-----KKTLNDA 259
>BG662253
Length = 342
Score = 81.3 bits (199), Expect = 2e-16
Identities = 43/103 (41%), Positives = 62/103 (59%)
Frame = +2
Query: 78 RWILDPADRYAVLANVAIKSINKDYHVIVEIASVLQPQELLAVRHAYHNRYKNSLEEDVA 137
R + P +R A L A+K Y V++EIA +ELL R AYH+ + +S+EEDVA
Sbjct: 11 RGTMHPWERDARLVKEALKKGPNAYGVLIEIACTRSSEELLGARKAYHSLFDHSIEEDVA 190
Query: 138 AHTSGYHRQLLVGLVSSFRYDGVEINPILAKHEADILHEAVKN 180
+H G R+LLV LVS++RY+ ++ AK EA + A+KN
Sbjct: 191 SHIHGIERKLLVALVSAYRYERSKVKDDTAKSEAKTISNAIKN 319
>BP073675
Length = 402
Score = 67.8 bits (164), Expect = 2e-12
Identities = 34/71 (47%), Positives = 45/71 (62%)
Frame = -3
Query: 244 YYEKVLRGALKRIGTDEDGLTRVVITRAEKDLKDIKELYYKRNSVHLEDTVAKEISGDYK 303
Y+ KVL A+K +GTD+ L RV++ R E DL +IK Y K+ L D V E SG+Y+
Sbjct: 400 YFAKVLHKAMKGLGTDDTKLIRVIVIRTEIDLHNIKAEYLKKYKKTLNDAVHSETSGNYR 221
Query: 304 KFLLTLLGKGH 314
FLL+LLG H
Sbjct: 220 AFLLSLLGPNH 188
Score = 32.7 bits (73), Expect = 0.082
Identities = 16/66 (24%), Positives = 35/66 (52%)
Frame = -3
Query: 87 YAVLANVAIKSINKDYHVIVEIASVLQPQELLAVRHAYHNRYKNSLEEDVAAHTSGYHRQ 146
+A + + A+K + D ++ + + +L ++ Y +YK +L + V + TSG +R
Sbjct: 397 FAKVLHKAMKGLGTDDTKLIRVIVIRTEIDLHNIKAEYLKKYKKTLNDAVHSETSGNYRA 218
Query: 147 LLVGLV 152
L+ L+
Sbjct: 217 FLLSLL 200
>CB829084
Length = 565
Score = 60.5 bits (145), Expect = 4e-10
Identities = 29/54 (53%), Positives = 39/54 (71%)
Frame = +2
Query: 143 YHRQLLVGLVSSFRYDGVEINPILAKHEADILHEAVKNKKGNIEEVIRILITRS 196
+ QLLV LV +FRYDG E+N ILAK EA +LHE + K N E+++R++ TRS
Sbjct: 404 FQYQLLVPLVGTFRYDGDEVNMILAKSEAKLLHEKIAEKAYNHEDLLRVITTRS 565
Score = 30.0 bits (66), Expect = 0.53
Identities = 17/32 (53%), Positives = 21/32 (65%)
Frame = +3
Query: 127 RYKNSLEEDVAAHTSGYHRQLLVGLVSSFRYD 158
R+K S+EEDVA HTSG R+ V L S+ D
Sbjct: 3 RFKRSIEEDVAYHTSGDIRK--VALTSNIIVD 92
>BP057141
Length = 466
Score = 60.1 bits (144), Expect = 5e-10
Identities = 27/48 (56%), Positives = 37/48 (76%)
Frame = +2
Query: 23 VKGWGTDESAIIAIMGQRNAVQRQQIRQAYQDIYQEDLIKRLESELSG 70
+KG G DE + AI+G R+ QRQQIR+ Y+++Y+E +IK LESELSG
Sbjct: 323 LKGCGADEKTVRAILGHRSVHQRQQIRKVYEELYREGIIKHLESELSG 466
>TC14375 similar to UP|O65848 (O65848) Annexin, partial (16%)
Length = 635
Score = 35.4 bits (80), Expect(2) = 0.002
Identities = 19/35 (54%), Positives = 23/35 (65%)
Frame = +1
Query: 148 LVGLVSSFRYDGVEINPILAKHEADILHEAVKNKK 182
LVGLV+SF Y G IN LA+ EA+ILH +K
Sbjct: 286 LVGLVASFTYIGEVINEKLAQTEAEILHRGPWRRK 390
Score = 21.9 bits (45), Expect(2) = 0.002
Identities = 11/16 (68%), Positives = 12/16 (74%)
Frame = +3
Query: 181 KKGNIEEVIRILITRS 196
KKG+ E IRIL TRS
Sbjct: 384 KKGSHGESIRILTTRS 431
>TC14376 similar to UP|O65848 (O65848) Annexin, partial (14%)
Length = 475
Score = 33.9 bits (76), Expect(2) = 0.005
Identities = 18/28 (64%), Positives = 20/28 (71%)
Frame = +1
Query: 148 LVGLVSSFRYDGVEINPILAKHEADILH 175
LVGLV+SF Y G IN LA EA+ILH
Sbjct: 121 LVGLVASFTYVGEVINEKLAHTEAEILH 204
Score = 21.9 bits (45), Expect(2) = 0.005
Identities = 11/16 (68%), Positives = 12/16 (74%)
Frame = +3
Query: 181 KKGNIEEVIRILITRS 196
KKG+ E IRIL TRS
Sbjct: 219 KKGSHGESIRILTTRS 266
>TC14374 similar to UP|O65848 (O65848) Annexin, partial (5%)
Length = 611
Score = 34.7 bits (78), Expect = 0.022
Identities = 22/53 (41%), Positives = 30/53 (56%)
Frame = +3
Query: 148 LVGLVSSFRYDGVEINPILAKHEADILHEAVKNKKGNIEEVIRILITRSKTQL 200
LVGLV+SF Y G IN L + EA+ILH +K + + + +KTQL
Sbjct: 204 LVGLVASFTYVGEVINEKLGQTEAEILH*GPWRRKEAMGSPLGS*LQGAKTQL 362
>AV776328
Length = 437
Score = 32.7 bits (73), Expect = 0.082
Identities = 21/50 (42%), Positives = 28/50 (56%)
Frame = -1
Query: 148 LVGLVSSFRYDGVEINPILAKHEADILHEAVKNKKGNIEEVIRILITRSK 197
L+GLV+SF Y G IN + + + KKG+ E IRIL TRS+
Sbjct: 395 LLGLVASFTYVGEVINEKVGSNRS*NPSLRYLEKKGSPGESIRILTTRSR 246
>TC14012 similar to UP|Q8GSM9 (Q8GSM9) Squalene monooxygenase 2 , partial
(13%)
Length = 480
Score = 27.3 bits (59), Expect = 3.4
Identities = 13/38 (34%), Positives = 18/38 (47%)
Frame = -3
Query: 122 HAYHNRYKNSLEEDVAAHTSGYHRQLLVGLVSSFRYDG 159
HA S EED H +HR++ +G +S R G
Sbjct: 199 HAVQREEAQSGEEDPTEHARIHHREMRIGRISPERRGG 86
>TC8211 homologue to UP|PDX1_PHAVU (Q9FT25) Probable pyridoxin biosynthesis
protein PDX1 (pvPDX1), partial (24%)
Length = 602
Score = 26.2 bits (56), Expect = 7.7
Identities = 11/36 (30%), Positives = 20/36 (55%)
Frame = -2
Query: 92 NVAIKSINKDYHVIVEIASVLQPQELLAVRHAYHNR 127
N+A+ I ++ + A++ Q + VRH+ HNR
Sbjct: 202 NIAVIQIEPNHSLPQSAANLRQQPRVTVVRHSLHNR 95
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.318 0.135 0.382
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,313,488
Number of Sequences: 28460
Number of extensions: 45322
Number of successful extensions: 211
Number of sequences better than 10.0: 42
Number of HSP's better than 10.0 without gapping: 205
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 208
length of query: 314
length of database: 4,897,600
effective HSP length: 90
effective length of query: 224
effective length of database: 2,336,200
effective search space: 523308800
effective search space used: 523308800
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 55 (25.8 bits)
Medicago: description of AC146590.13


