

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146588.4 - phase: 0
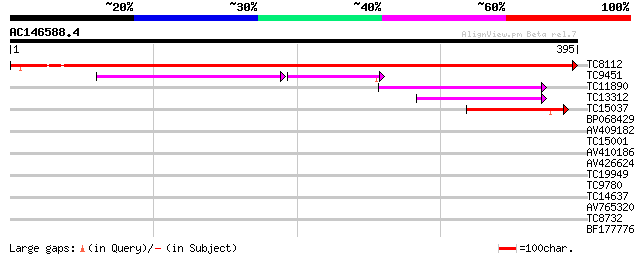
(395 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC8112 similar to UP|ODPA_PEA (P52902) Pyruvate dehydrogenase E1... 654 0.0
TC9451 homologue to UP|O24457 (O24457) Pyruvate dehydrogenase E1... 103 2e-36
TC11890 similar to PIR|A86347|A86347 branched-chain alpha keto-a... 74 3e-14
TC13312 weakly similar to PIR|A86347|A86347 branched-chain alpha... 54 5e-08
TC15037 similar to UP|O24457 (O24457) Pyruvate dehydrogenase E1 ... 52 2e-07
BP068429 38 0.003
AV409182 30 0.70
TC15001 similar to UP|GLE1_YEAST (Q12315) RNA export factor GLE1... 29 1.2
AV410186 29 1.6
AV426624 27 4.5
TC19949 UP|Q8H0D4 (Q8H0D4) Rac GTPase, partial (33%) 27 4.5
TC9780 similar to AAQ56830 (AAQ56830) At5g09670, partial (55%) 27 5.9
TC14637 weakly similar to UP|FUS_HUMAN (P35637) RNA-binding prot... 27 5.9
AV765320 27 7.7
TC8732 similar to UP|SNA2_ARATH (Q9SPE6) Alpha-soluble NSF attac... 27 7.7
BF177776 27 7.7
>TC8112 similar to UP|ODPA_PEA (P52902) Pyruvate dehydrogenase E1 component
alpha subunit, mitochondrial precursor (PDHE1-A) ,
partial (90%)
Length = 1816
Score = 654 bits (1688), Expect = 0.0
Identities = 320/401 (79%), Positives = 354/401 (87%), Gaps = 6/401 (1%)
Frame = +3
Query: 1 MALSRF------SSSQFGSTLLKPYFLSSAIRHRSISSSSTETLTVETSIPFTSHNCEPP 54
MALSR SSS GS LKP + ++R RSISS S +T+ETS+PFT+HNC+PP
Sbjct: 117 MALSRLASSSSSSSSTVGSKFLKPLTAAFSLR-RSISSDSNP-ITIETSVPFTAHNCDPP 290
Query: 55 STTVQTTASELMSFFNDMVLMRRMEIAADSLYKAKLIRGFCHLYDGQEAVAVGMEAAINR 114
S +V+T+ +EL SFF M MRRMEIAADSLYKAKLIRGFCHLYDGQEAVA+GMEA I +
Sbjct: 291 SRSVETSTAELHSFFRQMATMRRMEIAADSLYKAKLIRGFCHLYDGQEAVAIGMEAGITK 470
Query: 115 KDCVITAYRDHCTFLCRGGTLVEVFSELMGRKDGCSKGKGGSMHFYRKEGGFYGGHGIVG 174
KD +ITAYRDHCTFL RGGTL+E+FSELMGR DGCSKGKGGSMHFYRKE FYGGHGIVG
Sbjct: 471 KDAIITAYRDHCTFLGRGGTLLEIFSELMGRVDGCSKGKGGSMHFYRKETNFYGGHGIVG 650
Query: 175 AQIPLGVGLAFGQKYNKDPNVTFTLYGDGAANQGQLFEALNIAALWDLPAILVCENNHYG 234
AQ+PLG G+AF QKY+KD VTF LYGDGAANQGQLFEALNIAALWDLPAILVCENNHYG
Sbjct: 651 AQVPLGCGVAFAQKYSKDEAVTFALYGDGAANQGQLFEALNIAALWDLPAILVCENNHYG 830
Query: 235 MGTAEWRSAKSPAYYKRGDYAPGLKVDGMDVLAVKQACKFAKEHALKNGPLILEMDTYRY 294
MGTAEWR+AKSPAYYKRGDY PGLKVDGMDVLAVKQACKFAKEHALKNGP+ILEMDTYRY
Sbjct: 831 MGTAEWRAAKSPAYYKRGDYVPGLKVDGMDVLAVKQACKFAKEHALKNGPIILEMDTYRY 1010
Query: 295 HGHSMSDPGSTYRTRDEISGVRQERDPIERVRKLVLAHDISTEKELKDIEKEARKQVDEA 354
HGHSMSDPGSTYRTRDEISGVRQERDP+ERVRKL+++H+++TEKELKD+EKE RK+VDEA
Sbjct: 1011HGHSMSDPGSTYRTRDEISGVRQERDPLERVRKLLVSHEVATEKELKDMEKEVRKEVDEA 1190
Query: 355 IAKAKESQMPDPSDLFTNVYVKGLGVEACGADRKEVKATLP 395
IAKAKES MPDPSDL+ N+YVKG GVEA G DRKEV+ TLP
Sbjct: 1191IAKAKESSMPDPSDLYKNIYVKGYGVEAFGVDRKEVRVTLP 1313
>TC9451 homologue to UP|O24457 (O24457) Pyruvate dehydrogenase E1 alpha
subunit (At1g01090/T25K16_8) , partial (52%)
Length = 1082
Score = 103 bits (257), Expect(2) = 2e-36
Identities = 51/132 (38%), Positives = 72/132 (53%)
Frame = +1
Query: 61 TASELMSFFNDMVLMRRMEIAADSLYKAKLIRGFCHLYDGQEAVAVGMEAAINRKDCVIT 120
T E + + DM+L R E +Y + GF HLY+GQEAV+ G + ++D V++
Sbjct: 451 TKEEGLELYEDMILGRFFEDKCAEMYYRGKMFGFVHLYNGQEAVSTGFIKLLKKEDSVVS 630
Query: 121 AYRDHCTFLCRGGTLVEVFSELMGRKDGCSKGKGGSMHFYRKEGGFYGGHGIVGAQIPLG 180
YRDH L +G V SEL G+ G +G+GGSMH + KE GG +G IP+
Sbjct: 631 TYRDHVHALSKGVPARAVMSELFGKATGVCRGQGGSMHMFSKEHNVLGGFAFIGEGIPVA 810
Query: 181 VGLAFGQKYNKD 192
G AF KY ++
Sbjct: 811 TGAAFSSKYRRE 846
Score = 65.5 bits (158), Expect(2) = 2e-36
Identities = 30/70 (42%), Positives = 42/70 (59%), Gaps = 2/70 (2%)
Frame = +2
Query: 194 NVTFTLYGDGAANQGQLFEALNIAALWDLPAILVCENNHYGMGTAEWRSAKSPAYYKRGD 253
+VT +GDG N GQ +E LN+AALW LP + V ENN + +G + R+ P +K+G
Sbjct: 872 HVTLAFFGDGTCNNGQFYECLNMAALWKLPIVFVVENNLWAIGMSHLRATSDPEIWKKGP 1051
Query: 254 Y--APGLKVD 261
PG+ VD
Sbjct: 1052AFGMPGVHVD 1081
>TC11890 similar to PIR|A86347|A86347 branched-chain alpha keto-acid
dehydrogenase E1-alpha subunit [imported]
- Arabidopsis thaliana {Arabidopsis thaliana;}, partial
(32%)
Length = 621
Score = 74.3 bits (181), Expect = 3e-14
Identities = 43/118 (36%), Positives = 68/118 (57%), Gaps = 1/118 (0%)
Frame = +1
Query: 258 LKVDGMDVLAVKQACKFAKEHALKNG-PLILEMDTYRYHGHSMSDPGSTYRTRDEISGVR 316
++VDG D LAV A A+E A++ P+++E TYR HS SD + YR DEI +
Sbjct: 22 IRVDGNDALAVYSAVHTAREIAIREQRPVLIEALTYRVGHHSTSDDSTKYRPVDEIEYWK 201
Query: 317 QERDPIERVRKLVLAHDISTEKELKDIEKEARKQVDEAIAKAKESQMPDPSDLFTNVY 374
R+P+ R ++ V + ++K+ ++ RKQ+ AI A+++Q P DLF +VY
Sbjct: 202 MARNPVNRFKRWVEMNGWWSDKDELELRSSVRKQLMNAIQVAEKAQKPPLEDLFHDVY 375
>TC13312 weakly similar to PIR|A86347|A86347 branched-chain alpha keto-acid
dehydrogenase E1-alpha subunit [imported]
- Arabidopsis thaliana {Arabidopsis thaliana;}, partial
(25%)
Length = 510
Score = 53.9 bits (128), Expect = 5e-08
Identities = 29/91 (31%), Positives = 46/91 (49%)
Frame = +2
Query: 284 PLILEMDTYRYHGHSMSDPGSTYRTRDEISGVRQERDPIERVRKLVLAHDISTEKELKDI 343
P+++E TYR HS SD + YR EI RQ RDP+ + RK + + ++
Sbjct: 20 PVLIEALTYRVGHHSTSDDSTKYRPASEIEWWRQARDPVAKFRKWIERNGWWNAVAESEL 199
Query: 344 EKEARKQVDEAIAKAKESQMPDPSDLFTNVY 374
R+Q+ I A+ + P D+F++VY
Sbjct: 200 RNSLRQQLLHTIQVAESVEKPPLGDIFSDVY 292
>TC15037 similar to UP|O24457 (O24457) Pyruvate dehydrogenase E1 alpha
subunit (At1g01090/T25K16_8) , partial (20%)
Length = 499
Score = 51.6 bits (122), Expect = 2e-07
Identities = 28/73 (38%), Positives = 45/73 (61%), Gaps = 2/73 (2%)
Frame = +2
Query: 319 RDPIERVRKLVLAHDISTEKELKDIEKEARKQVDEAIAKAKESQMPDPSDLFTNVYV--K 376
RDPI ++K + +++++E+ELK IEK+ + ++EA+ A ES +P S L NV+ K
Sbjct: 5 RDPITALKKYIFENNLASEQELKAIEKKIDEVLEEAVEFADESPLPPRSQLLENVFADPK 184
Query: 377 GLGVEACGADRKE 389
G G+ G R E
Sbjct: 185 GFGIGPDGKYRCE 223
>BP068429
Length = 412
Score = 38.1 bits (87), Expect = 0.003
Identities = 22/60 (36%), Positives = 36/60 (59%), Gaps = 2/60 (3%)
Frame = -2
Query: 332 HDISTEKELKDIEKEARKQVDEAIAKAKESQMPDPSDLFTNVYV--KGLGVEACGADRKE 389
+++++ +ELK IEK+ + ++EA+ A ES +P PS L NV+ K G+ G R E
Sbjct: 411 NNLASAQELKAIEKKIDEVLEEAVDFADESPLPPPSQLLENVFADPKPFGIGPDGKYRCE 232
>AV409182
Length = 312
Score = 30.0 bits (66), Expect = 0.70
Identities = 21/64 (32%), Positives = 32/64 (49%), Gaps = 11/64 (17%)
Frame = +1
Query: 180 GVGLAFGQK-----YNKDPN-----VTFTLYGDGAANQGQLFEALNIAALWDL-PAILVC 228
GVGLA +K +NK + T+ + GDG +G EA ++A W L I +
Sbjct: 16 GVGLALAEKHLAARFNKPDSEIVDHYTYVILGDGCQMEGISNEACSLAGHWGLGKLIALY 195
Query: 229 ENNH 232
++NH
Sbjct: 196 DDNH 207
>TC15001 similar to UP|GLE1_YEAST (Q12315) RNA export factor GLE1, partial
(4%)
Length = 988
Score = 29.3 bits (64), Expect = 1.2
Identities = 16/36 (44%), Positives = 21/36 (57%)
Frame = +2
Query: 328 LVLAHDISTEKELKDIEKEARKQVDEAIAKAKESQM 363
L+ H E ELK IE+E K+V+EAI K E +
Sbjct: 341 LLCFHRRRKEAELKLIEEETAKRVEEAIRKRVEESL 448
>AV410186
Length = 433
Score = 28.9 bits (63), Expect = 1.6
Identities = 22/66 (33%), Positives = 30/66 (45%), Gaps = 14/66 (21%)
Frame = +2
Query: 22 SSAIRHRSISSSSTETLTVETSI--------------PFTSHNCEPPSTTVQTTASELMS 67
SS+I SISSSS+ + E S+ P SH+ P+TT T +E S
Sbjct: 44 SSSILQLSISSSSSSSPETEPSLRNAH*SHLHLPRTFPLISHHPSAPNTTHLATITESSS 223
Query: 68 FFNDMV 73
F +V
Sbjct: 224 FLCPLV 241
>AV426624
Length = 422
Score = 27.3 bits (59), Expect = 4.5
Identities = 18/55 (32%), Positives = 26/55 (46%), Gaps = 4/55 (7%)
Frame = -2
Query: 4 SRFSSSQFGSTLLKPYFLSSAIRHRSISSSSTETLTVET----SIPFTSHNCEPP 54
S SSS S+L +P++ SS ++ + + TL T S P NC PP
Sbjct: 379 SSSSSSSSSSSL*EPFYSSSHLKKQKPQIFNLRTLMTHTQTRYSHPVRFPNCAPP 215
>TC19949 UP|Q8H0D4 (Q8H0D4) Rac GTPase, partial (33%)
Length = 549
Score = 27.3 bits (59), Expect = 4.5
Identities = 18/55 (32%), Positives = 26/55 (46%), Gaps = 4/55 (7%)
Frame = -3
Query: 4 SRFSSSQFGSTLLKPYFLSSAIRHRSISSSSTETLTVET----SIPFTSHNCEPP 54
S SSS S+L +P++ SS ++ + + TL T S P NC PP
Sbjct: 352 SSSSSSSSSSSL*EPFYSSSHLKKQKPQIFNLRTLMTHTQTRYSHPVRFPNCAPP 188
>TC9780 similar to AAQ56830 (AAQ56830) At5g09670, partial (55%)
Length = 1250
Score = 26.9 bits (58), Expect = 5.9
Identities = 14/34 (41%), Positives = 16/34 (46%), Gaps = 4/34 (11%)
Frame = +2
Query: 148 GCSKGKGGSMHFYRKEGG----FYGGHGIVGAQI 177
GCSKG GS + GG Y G GI G +
Sbjct: 920 GCSKGAEGSTPLCKAHGGGKRCLYNGGGICGKSV 1021
>TC14637 weakly similar to UP|FUS_HUMAN (P35637) RNA-binding protein FUS
(Oncogene FUS) (Oncogene TLS) (Translocated in
liposarcoma protein) (POMp75) (75 kDa DNA-pairing
protein), partial (6%)
Length = 1332
Score = 26.9 bits (58), Expect = 5.9
Identities = 14/38 (36%), Positives = 17/38 (43%)
Frame = +2
Query: 137 EVFSELMGRKDGCSKGKGGSMHFYRKEGGFYGGHGIVG 174
E F + + G G+GG R GGFYG G G
Sbjct: 857 ETFGDSARYRGGGRGGRGGPFRGGRTRGGFYGRGGGFG 970
>AV765320
Length = 572
Score = 26.6 bits (57), Expect = 7.7
Identities = 19/63 (30%), Positives = 31/63 (49%), Gaps = 2/63 (3%)
Frame = +1
Query: 7 SSSQFGSTLLKPYF--LSSAIRHRSISSSSTETLTVETSIPFTSHNCEPPSTTVQTTASE 64
+++ G +L P+ +SS+ SI SSST V++ F PP+ T +T S
Sbjct: 340 ATTSTGRMILTPFLAAISSSSLASSILSSSTSEEPVDSPRAFRKVKTMPPTITSLSTLSS 519
Query: 65 LMS 67
+ S
Sbjct: 520 IDS 528
>TC8732 similar to UP|SNA2_ARATH (Q9SPE6) Alpha-soluble NSF attachment
protein 2 (Alpha-SNAP2) (N-ethylmaleimide-sensitive
factor attachment protein, alpha 2), complete
Length = 1490
Score = 26.6 bits (57), Expect = 7.7
Identities = 14/47 (29%), Positives = 24/47 (50%)
Frame = -3
Query: 330 LAHDISTEKELKDIEKEARKQVDEAIAKAKESQMPDPSDLFTNVYVK 376
L+HD+++ KEL D+ K + A + E P P F+ ++ K
Sbjct: 396 LSHDLASLKELADLSKSS------AASSNLEPNKPQPLSFFSALFKK 274
>BF177776
Length = 384
Score = 26.6 bits (57), Expect = 7.7
Identities = 14/33 (42%), Positives = 17/33 (51%)
Frame = -2
Query: 33 SSTETLTVETSIPFTSHNCEPPSTTVQTTASEL 65
SS + T +S+P S PP T QT AS L
Sbjct: 248 SSPPSPTCNSSLPSLSSPASPPETPPQTAASTL 150
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.318 0.135 0.396
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,499,861
Number of Sequences: 28460
Number of extensions: 88799
Number of successful extensions: 465
Number of sequences better than 10.0: 32
Number of HSP's better than 10.0 without gapping: 456
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 461
length of query: 395
length of database: 4,897,600
effective HSP length: 92
effective length of query: 303
effective length of database: 2,279,280
effective search space: 690621840
effective search space used: 690621840
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 56 (26.2 bits)
Medicago: description of AC146588.4


