

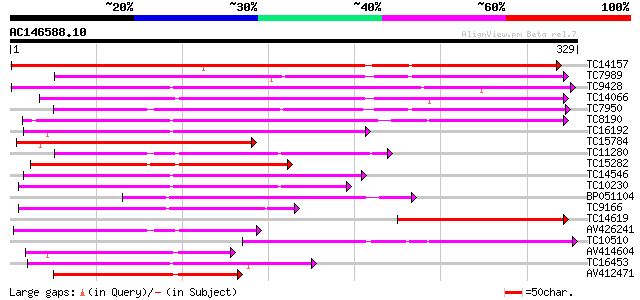
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146588.10 - phase: 0
(329 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC14157 similar to UP|O24080 (O24080) Peroxidase2 precursor , p... 257 2e-69
TC7989 similar to UP|Q43854 (Q43854) Peroxidase precursor , par... 204 2e-53
TC9428 similar to UP|Q9XFL3 (Q9XFL3) Peroxidase 1 (Fragment), pa... 202 5e-53
TC14066 homologue to UP|O64970 (O64970) Cationic peroxidase 2 ,... 196 3e-51
TC7950 similar to UP|Q9XFI8 (Q9XFI8) Peroxidase (Fragment), part... 196 4e-51
TC8190 similar to UP|PER1_ARAHY (P22195) Cationic peroxidase 1 p... 184 2e-47
TC16192 similar to UP|Q9SSZ9 (Q9SSZ9) Peroxidase 1, partial (49%) 166 6e-42
TC15784 similar to UP|PE31_ARATH (Q9LHA7) Peroxidase 31 precurso... 150 4e-37
TC11280 similar to UP|Q9XFL5 (Q9XFL5) Peroxidase 4 (Fragment), p... 150 4e-37
TC15282 similar to UP|Q9ZNZ6 (Q9ZNZ6) Peroxidase precursor (Fra... 137 2e-33
TC14546 similar to UP|Q9XGV6 (Q9XGV6) Bacterial-induced peroxida... 137 2e-33
TC10230 similar to UP|PE10_ARATH (Q9FX85) Peroxidase 10 precurso... 137 2e-33
BP051104 127 2e-30
TC9166 similar to UP|Q9XFL4 (Q9XFL4) Peroxidase 3, partial (49%) 119 7e-28
TC14619 similar to GB|CAA72487.1|1890317|ATP26A peroxidase ATP26... 115 1e-26
AV426241 114 3e-26
TC10510 similar to PIR|B56555|B56555 peroxidase , anionic, precu... 107 4e-24
AV414604 106 6e-24
TC16453 similar to UP|Q40372 (Q40372) Peroxidase precursor, part... 105 1e-23
AV412471 102 9e-23
>TC14157 similar to UP|O24080 (O24080) Peroxidase2 precursor , partial
(96%)
Length = 1390
Score = 257 bits (656), Expect = 2e-69
Identities = 137/321 (42%), Positives = 196/321 (60%), Gaps = 2/321 (0%)
Frame = +1
Query: 2 IYKSYIFSLLCLALFPITQSKLIPNYYQKSCPKFEDIVKQTVTDKQKTTPSTAGAALRLF 61
I + + SL F ++L PN+Y +CP + IVK V K + T T A LRLF
Sbjct: 64 INRVLVLSLSLTLCFYTCFAQLSPNHYASTCPNLQSIVKGVVQKKFQQTFVTVPATLRLF 243
Query: 62 FSDCMIGGCDASVLVSSNSFNKAERDADINLSLSGDGFEVVTRAKNMLEL--ECPGVVSC 119
F DC + GCDASV+V+S+ NKAE+D NLSL+GDGF+ V +AK ++ +C VSC
Sbjct: 244 FHDCFVQGCDASVMVASSGNNKAEKDHPDNLSLAGDGFDTVIKAKAAVDAVPQCRNKVSC 423
Query: 120 ADILAAAARDLVVSVGGPFYELDLGRRDSLESKSIDAENKYPLPTMTNSQVIDIFTSKGF 179
ADILA A RD+VV GGP Y ++LGR D L S++ D + P P +Q+ +F S+G
Sbjct: 424 ADILALATRDVVVLAGGPSYTVELGRFDGLVSRASDVNGRLPEPNFNLNQLNSLFASQGL 603
Query: 180 TVQEMVALAGAHTIGFSHCKQFSNRLFNFSKTTETDPKYNPEYAAGLKKLCQNYQKDTSM 239
T +M+AL+GAHT+GFSHC +FSNR++ +T DP N YA L+++C + +
Sbjct: 604 TQTDMIALSGAHTLGFSHCNRFSNRIY----STPVDPTLNRNYATQLQQMCPK-NVNPQI 768
Query: 240 SAFNDVMTPSKFDNMYFKNLKRGMGLLATDSLMGEDKRTKPFVDMYAENQTKFFEDFGNA 299
+ D TP FDN+Y+KNL++G GL +D ++ D+R+K V+ +A N F +F A
Sbjct: 769 AINMDPTTPRTFDNIYYKNLQQGKGLFTSDQILFTDQRSKATVNSFASNSNTFNANFAAA 948
Query: 300 MRKLSVLHVKEGKDGEIRNRC 320
M KL + VK ++G+IR C
Sbjct: 949 MIKLGRVGVKTARNGKIRTDC 1011
>TC7989 similar to UP|Q43854 (Q43854) Peroxidase precursor , partial (85%)
Length = 1352
Score = 204 bits (518), Expect = 2e-53
Identities = 113/300 (37%), Positives = 165/300 (54%), Gaps = 2/300 (0%)
Frame = +3
Query: 27 YYQKSCPKFEDIVKQTVTDKQKTTPSTAGAALRLFFSDCMIGGCDASVLVSSNSFNKAER 86
+Y SCPK E I+++ + + A LRL F DC + GCDASVL+ ++ +E+
Sbjct: 150 FYDSSCPKLESIIRKELKKVFDKDIAQAAGLLRLHFHDCFVQGCDASVLLDGSASGPSEK 329
Query: 87 DADINLSLSGDGFEVVTRAKNMLELECPGVVSCADILAAAARDLVVSVGGPFYELDLGRR 146
DA NL+L F+++ ++ +E +C VVSCADI A AARD V GGP YEL LGRR
Sbjct: 330 DAPPNLTLRAQAFKIIEDLRSQIEKKCGRVVSCADITAIAARDAVFLSGGPDYELPLGRR 509
Query: 147 DSLE--SKSIDAENKYPLPTMTNSQVIDIFTSKGFTVQEMVALAGAHTIGFSHCKQFSNR 204
D L ++++ +N P P + +++ +K ++V+L+G HTIG SHC F++R
Sbjct: 510 DGLNFATRNVTLDN-LPAPQSNTTTILNSLATKNLDPTDVVSLSGGHTIGISHCSSFTDR 686
Query: 205 LFNFSKTTETDPKYNPEYAAGLKKLCQNYQKDTSMSAFNDVMTPSKFDNMYFKNLKRGMG 264
L+ DP + + LK C +T + D+ +P+ FDN Y+ +L G
Sbjct: 687 LY-----PSKDPVMDQTFEKNLKLTCP--ASNTDNTTVLDLRSPNTFDNKYYVDLMNRQG 845
Query: 265 LLATDSLMGEDKRTKPFVDMYAENQTKFFEDFGNAMRKLSVLHVKEGKDGEIRNRCDTFN 324
L +D + DKRTK V +A NQ+ FFE F AM K+ L+V G GEIR C N
Sbjct: 846 LFFSDQDLYTDKRTKDIVTSFAVNQSLFFEKFVVAMLKMGQLNVLTGSQGEIRANCSVRN 1025
>TC9428 similar to UP|Q9XFL3 (Q9XFL3) Peroxidase 1 (Fragment), partial
(97%)
Length = 1354
Score = 202 bits (515), Expect = 5e-53
Identities = 121/330 (36%), Positives = 178/330 (53%), Gaps = 3/330 (0%)
Frame = +2
Query: 2 IYKSYIFSLLCLALFPITQSKLIPN-YYQKSCPKFEDIVKQTVTDKQKTTPSTAGAALRL 60
I+ + +F ++ AL P + + N +Y+ +CP IV++ V + K+ P G+ +RL
Sbjct: 50 IHLAVVFCVVIGALVPYSSDAQLDNSFYKDTCPNVHSIVREVVRNVSKSDPRMLGSLIRL 229
Query: 61 FFSDCMIGGCDASVLVSSNSFNKAERDADINLSLSGDGFEVVTRAKNMLELECPGVVSCA 120
F DC + GCDAS+L++ S +E+ A N + S G +VV + K +E CP VSCA
Sbjct: 230 HFHDCFVQGCDASILLNDTSTIVSEQGAFPNNN-SIRGLDVVNQIKTAVENACPNTVSCA 406
Query: 121 DILAAAARDLVVSVGGPFYELDLGRRDSLESKSIDAENKYPLPTMTNSQVIDIFTSKGFT 180
DILA AA V G +++ LGRRDSL + A P P SQ+ F +
Sbjct: 407 DILALAAEISSVLANGVDWKVPLGRRDSLTANRSLANQNLPGPNFNLSQLNASFAVQNLN 586
Query: 181 VQEMVALAGAHTIGFSHCKQFSNRLFNFSKTTETDPKYNPEYAAGLKKLCQNYQKDTSMS 240
+ ++VAL+GAHTIG C+ F NRL+NFS T DP N Y L+ +C + T++
Sbjct: 587 ITDLVALSGAHTIGRGQCQFFVNRLYNFSNTGNPDPTLNTTYLQTLRSICPHGGPGTTL- 763
Query: 241 AFNDVMTPSKFDNMYFKNLKRGMGLLATDSLM--GEDKRTKPFVDMYAENQTKFFEDFGN 298
A D TP D+ YF NL+ G GL +D ++ T V+ ++ N T FFE+F
Sbjct: 764 AHLDPTTPDTLDSQYFSNLQGGKGLFQSDPVLFSTSGAATVAIVNSFSSNPTLFFENFKA 943
Query: 299 AMRKLSVLHVKEGKDGEIRNRCDTFNNLNA 328
+M K+S + V G GEIR C+ N +A
Sbjct: 944 SMIKMSRIGVLTGSQGEIRKPCNFVNGNSA 1033
>TC14066 homologue to UP|O64970 (O64970) Cationic peroxidase 2 , partial
(92%)
Length = 1436
Score = 196 bits (499), Expect = 3e-51
Identities = 113/309 (36%), Positives = 167/309 (53%), Gaps = 2/309 (0%)
Frame = +2
Query: 18 ITQSKLIPNYYQKSCPKFEDIVKQTVTDKQKTTPSTAGAALRLFFSDCMIGGCDASVLVS 77
+ + L N+Y+++CP+ EDI+ + V K +TA + LR F DC + CDAS+L+
Sbjct: 137 VQDTGLAMNFYKETCPQAEDIITEQVKLLYKRHKNTAFSWLRNIFHDCAVQSCDASLLLD 316
Query: 78 SNSFNKAERDADINLSLSGDGFEVVTRAKNMLELECPGVVSCADILAAAARDLVVSVGGP 137
S +E++ D + L F + K LE ECPGVVSC+DIL +ARD + ++GGP
Sbjct: 317 STRKTLSEKETDRSFGLRN--FRYIETIKEALERECPGVVSCSDILVLSARDGIAALGGP 490
Query: 138 FYELDLGRRDSLESKSIDAENKYPLPTMTNSQVIDIFTSKGFTVQEMVALAGAHTIGFSH 197
L GRRD S++ E P + S V+D F + G +VAL GAH++G +H
Sbjct: 491 HIPLRTGRRDGRRSRADVVEQFLPDHNESISAVLDKFGAMGIDTPGVVALLGAHSVGRTH 670
Query: 198 CKQFSNRLFNFSKTTETDPKYNPEYAAGLKKLCQNYQKDTSMSAF--NDVMTPSKFDNMY 255
C + +RL+ E DP NP++ + K C + D + ND TP DN Y
Sbjct: 671 CVKLVHRLY-----PEVDPALNPDHIPHMLKKCPDAIPDPKAVQYVRNDRGTPMILDNNY 835
Query: 256 FKNLKRGMGLLATDSLMGEDKRTKPFVDMYAENQTKFFEDFGNAMRKLSVLHVKEGKDGE 315
++N+ GLL D + DKRTKP+V A++Q FF++F A+ LS + G GE
Sbjct: 836 YRNILDNKGLLLVDHQLANDKRTKPYVKKMAKSQDYFFKEFSRAITLLSENNPLTGTKGE 1015
Query: 316 IRNRCDTFN 324
IR +C+ N
Sbjct: 1016IRKQCNVAN 1042
>TC7950 similar to UP|Q9XFI8 (Q9XFI8) Peroxidase (Fragment), partial (86%)
Length = 1286
Score = 196 bits (498), Expect = 4e-51
Identities = 108/300 (36%), Positives = 161/300 (53%)
Frame = +1
Query: 26 NYYQKSCPKFEDIVKQTVTDKQKTTPSTAGAALRLFFSDCMIGGCDASVLVSSNSFNKAE 85
++Y K+CPK E +V+ + K A LR+FF DC + GCD SVL+ + E
Sbjct: 115 SFYSKTCPKLETVVRNHLKKVLKKDNGQAPGLLRIFFHDCFVQGCDGSVLLDGSP---GE 285
Query: 86 RDADINLSLSGDGFEVVTRAKNMLELECPGVVSCADILAAAARDLVVSVGGPFYELDLGR 145
RD N+ + + + + + ++ +C +VSCADI A+RD V GGP Y + LGR
Sbjct: 286 RDQPANIGIRPEALQTIEDIRALVHKQCGKIVSCADITILASRDAVFLTGGPDYAVPLGR 465
Query: 146 RDSLESKSIDAENKYPLPTMTNSQVIDIFTSKGFTVQEMVALAGAHTIGFSHCKQFSNRL 205
RD + ++ + K P P + + F + F ++VAL+GAHT G +HC F NRL
Sbjct: 466 RDGVSFSTVGTQ-KLPSPINNTTATLKAFADRNFDATDVVALSGAHTFGRAHCGTFFNRL 642
Query: 206 FNFSKTTETDPKYNPEYAAGLKKLCQNYQKDTSMSAFNDVMTPSKFDNMYFKNLKRGMGL 265
+ DP + A L C ++++ +A D+ TP+ FDN Y+ +L G+
Sbjct: 643 ------SPLDPNMDKTLAKNLTATCP--AQNSTNTANLDIRTPNVFDNKYYLDLMNRQGV 798
Query: 266 LATDSLMGEDKRTKPFVDMYAENQTKFFEDFGNAMRKLSVLHVKEGKDGEIRNRCDTFNN 325
+D + DKRTK V+ +A NQT FFE F +A+ KLS L V G GEIR RC+ N+
Sbjct: 799 FTSDQDLLSDKRTKGLVNAFAVNQTLFFEKFVDAVIKLSQLDVLTGNQGEIRGRCNVVNS 978
>TC8190 similar to UP|PER1_ARAHY (P22195) Cationic peroxidase 1 precursor
(PNPC1) , partial (96%)
Length = 1202
Score = 184 bits (466), Expect = 2e-47
Identities = 114/317 (35%), Positives = 165/317 (51%)
Frame = +1
Query: 8 FSLLCLALFPITQSKLIPNYYQKSCPKFEDIVKQTVTDKQKTTPSTAGAALRLFFSDCMI 67
F L CL I ++L +Y K+CP +K V + LRL F DC +
Sbjct: 103 FFLFCL--IGIGSAQLSSTFYAKTCPLVLATIKTQVNLAVAKEARMGASLLRLHFHDCFV 276
Query: 68 GGCDASVLVSSNSFNKAERDADINLSLSGDGFEVVTRAKNMLELECPGVVSCADILAAAA 127
GCDAS+L+ S E+ A N + S G++V+ K+ +E CPGVVSCADI+A AA
Sbjct: 277 QGCDASILLDDTSSFTGEKTAGPNAN-SVRGYDVIDTIKSKVESLCPGVVSCADIVAVAA 453
Query: 128 RDLVVSVGGPFYELDLGRRDSLESKSIDAENKYPLPTMTNSQVIDIFTSKGFTVQEMVAL 187
RD VV++GG + + LGRRDS + A ++ P P+ + F++KGFT +EMVAL
Sbjct: 454 RDSVVALGGFSWAVPLGRRDSTTASLSSANSELPGPSSNLDGLNTAFSNKGFTTREMVAL 633
Query: 188 AGAHTIGFSHCKQFSNRLFNFSKTTETDPKYNPEYAAGLKKLCQNYQKDTSMSAFNDVMT 247
+G+HTIG + C F R++ + T +A L+ C D+++S D +
Sbjct: 634 SGSHTIGQARCLFFRTRIYYETHIDST-------FAKNLQGNCPFNGGDSNLSPL-DTTS 789
Query: 248 PSKFDNMYFKNLKRGMGLLATDSLMGEDKRTKPFVDMYAENQTKFFEDFGNAMRKLSVLH 307
P+ FDN Y++NL+ GL +D ++ T V+ Y N F DF NAM K+ L
Sbjct: 790 PTTFDNGYYRNLQSQKGLFHSDQVLFNGGSTDSQVNSYVTNPASFKTDFANAMVKMGNLS 969
Query: 308 VKEGKDGEIRNRCDTFN 324
G G+IR C N
Sbjct: 970 PLTGSSGQIRTHCRKTN 1020
>TC16192 similar to UP|Q9SSZ9 (Q9SSZ9) Peroxidase 1, partial (49%)
Length = 645
Score = 166 bits (419), Expect = 6e-42
Identities = 95/204 (46%), Positives = 118/204 (57%), Gaps = 3/204 (1%)
Frame = +2
Query: 9 SLLCLALFPITQ---SKLIPNYYQKSCPKFEDIVKQTVTDKQKTTPSTAGAALRLFFSDC 65
+++ L +F + Q S+L YY SC E IVK V P A +R+ F DC
Sbjct: 32 AIIVLVIFLLNQNAYSELEVGYYSYSCGMAEFIVKDEVRRSVTKNPGIAAGLVRMHFHDC 211
Query: 66 MIGGCDASVLVSSNSFNKAERDADINLSLSGDGFEVVTRAKNMLELECPGVVSCADILAA 125
I GCDASVL+ S N AE+D+ N S GFEV+ AK LE C GVVSCADI+A
Sbjct: 212 FIRGCDASVLLDSTPSNTAEKDSPANKP-SLRGFEVIDSAKAKLEAVCKGVVSCADIIAF 388
Query: 126 AARDLVVSVGGPFYELDLGRRDSLESKSIDAENKYPLPTMTNSQVIDIFTSKGFTVQEMV 185
AARD V GG Y++ GRRD S + D P PT +Q+ +F KG T +EMV
Sbjct: 389 AARDSVELAGGVGYDVPAGRRDGRISLASDTRTDLPPPTFNVNQLTQLFAKKGLTQEEMV 568
Query: 186 ALAGAHTIGFSHCKQFSNRLFNFS 209
L+GAHTIG SHC FS+RL+NFS
Sbjct: 569 TLSGAHTIGRSHCAAFSSRLYNFS 640
>TC15784 similar to UP|PE31_ARATH (Q9LHA7) Peroxidase 31 precursor (Atperox
P31) (ATP41) , partial (38%)
Length = 475
Score = 150 bits (378), Expect = 4e-37
Identities = 77/142 (54%), Positives = 100/142 (70%), Gaps = 3/142 (2%)
Frame = +3
Query: 5 SYIFSLLCLALF--PITQSKLIPNYYQKSCPKFEDIVKQTVTDKQKTTPSTAGAALRLFF 62
S +F LL F ++ ++L ++Y+ +CP+F I++ TVT+KQ TTP+T A LRLF
Sbjct: 48 SSLFLLLSTLSFFTTLSHARLTLDFYKHTCPQFSQIIRDTVTNKQITTPTTGPATLRLFL 227
Query: 63 SDCMI-GGCDASVLVSSNSFNKAERDADINLSLSGDGFEVVTRAKNMLELECPGVVSCAD 121
DC+ GCDAS+L+SS SFNKAERDADINLSL GD F++V RAK LEL CP VSCAD
Sbjct: 228 HDCLHPNGCDASILLSSTSFNKAERDADINLSLPGDAFDLVVRAKTALELACPNTVSCAD 407
Query: 122 ILAAAARDLVVSVGGPFYELDL 143
IL AA DL+ +GGP + + L
Sbjct: 408 ILTAATSDLLTMLGGPHFPVFL 473
>TC11280 similar to UP|Q9XFL5 (Q9XFL5) Peroxidase 4 (Fragment), partial
(68%)
Length = 614
Score = 150 bits (378), Expect = 4e-37
Identities = 87/196 (44%), Positives = 115/196 (58%)
Frame = +3
Query: 27 YYQKSCPKFEDIVKQTVTDKQKTTPSTAGAALRLFFSDCMIGGCDASVLVSSNSFNKAER 86
+Y +CP+ E IV+ TV + P+ A LR+ F DC + GCDASVL++ ER
Sbjct: 36 FYLGTCPRAESIVRSTVESHVNSDPTLAAGLLRMHFHDCFVQGCDASVLIAGAG---TER 206
Query: 87 DADINLSLSGDGFEVVTRAKNMLELECPGVVSCADILAAAARDLVVSVGGPFYELDLGRR 146
A NLSL G FEV+ AK +E CPGVVSCADILA AARD VV GG +++ GRR
Sbjct: 207 TAIPNLSLRG--FEVIDDAKAKVEAACPGVVSCADILALAARDSVVLSGGLSWQVPTGRR 380
Query: 147 DSLESKSIDAENKYPLPTMTNSQVIDIFTSKGFTVQEMVALAGAHTIGFSHCKQFSNRLF 206
D S++ D N P P + FT+KG Q++V L G HTIG + C+ FSNRL+
Sbjct: 381 DGRVSQASDV-NNLPAPFDSVDVQKQKFTAKGLNTQDLVTLVGGHTIGTTACQFFSNRLY 557
Query: 207 NFSKTTETDPKYNPEY 222
NF+ + DP + +
Sbjct: 558 NFT-SNGPDPSIDASF 602
>TC15282 similar to UP|Q9ZNZ6 (Q9ZNZ6) Peroxidase precursor (Fragment) ,
partial (47%)
Length = 604
Score = 137 bits (346), Expect = 2e-33
Identities = 69/152 (45%), Positives = 101/152 (66%)
Frame = +1
Query: 13 LALFPITQSKLIPNYYQKSCPKFEDIVKQTVTDKQKTTPSTAGAALRLFFSDCMIGGCDA 72
LAL ++L +Y K+CPK E I+ V + PS A A +R+ F DC + GCDA
Sbjct: 133 LALIASNHAQLEQGFYTKTCPKAEKIILDFVHEHIHNAPSLAAALIRMNFHDCFVRGCDA 312
Query: 73 SVLVSSNSFNKAERDADINLSLSGDGFEVVTRAKNMLELECPGVVSCADILAAAARDLVV 132
S+L++S S +AERDA NL++ G F+ + R K+++E +CPGVVSCADI+A AARD +V
Sbjct: 313 SILLNSTS-KQAERDAPPNLTVRG--FDFIDRIKSLVEAQCPGVVSCADIIALAARDSIV 483
Query: 133 SVGGPFYELDLGRRDSLESKSIDAENKYPLPT 164
+ GGPF+++ GRRD + S ++A ++ P PT
Sbjct: 484 ATGGPFWKVPTGRRDGVISNLVEARSQIPAPT 579
>TC14546 similar to UP|Q9XGV6 (Q9XGV6) Bacterial-induced peroxidase
precursor , partial (66%)
Length = 738
Score = 137 bits (346), Expect = 2e-33
Identities = 73/200 (36%), Positives = 113/200 (56%), Gaps = 1/200 (0%)
Frame = +1
Query: 9 SLLCLALFPITQSKLIPNYYQKSCPKFEDIVKQTVTDKQKTTPSTAGAALRLFFSDCMIG 68
S+L L ++L P +Y +SCPK + IV+ + K + LRLFF DC +
Sbjct: 58 SILLSLLACSANAQLTPTFYDRSCPKLQTIVRNAMVQTIKKEARMGASILRLFFHDCFVN 237
Query: 69 GCDASVLVSS-NSFNKAERDADINLSLSGDGFEVVTRAKNMLELECPGVVSCADILAAAA 127
GCD S+L+ + E++A N + S GFEV+ K +E C VSCADILA A
Sbjct: 238 GCDGSILLDDIGTTFVGEKNAAPNKN-SARGFEVIDTIKTNVEASCNNTVSCADILALAT 414
Query: 128 RDLVVSVGGPFYELDLGRRDSLESKSIDAENKYPLPTMTNSQVIDIFTSKGFTVQEMVAL 187
RD + +GGP +++ LGRRD+ + A + P P+ S +I +F++KG + +++ L
Sbjct: 415 RDGINLLGGPTWQVPLGRRDARTASQSKANTEIPSPSSDLSTLISMFSAKGLSARDLTVL 594
Query: 188 AGAHTIGFSHCKQFSNRLFN 207
+G HTIG + C+ F +R+ N
Sbjct: 595 SGGHTIGQAECQFFRSRVNN 654
>TC10230 similar to UP|PE10_ARATH (Q9FX85) Peroxidase 10 precursor (Atperox
P10) (ATP5a) , partial (42%)
Length = 753
Score = 137 bits (345), Expect = 2e-33
Identities = 75/193 (38%), Positives = 103/193 (52%)
Frame = +3
Query: 6 YIFSLLCLALFPITQSKLIPNYYQKSCPKFEDIVKQTVTDKQKTTPSTAGAALRLFFSDC 65
++F L + L P+ S+L N+Y +CP IV+ + A + LRL F DC
Sbjct: 174 FVFMLWLVLLSPLVSSQLYYNFYDTTCPNLTRIVRYNLLSAMSNDTRIAASLLRLHFHDC 353
Query: 66 MIGGCDASVLVSSNSFNKAERDADINLSLSGDGFEVVTRAKNMLELECPGVVSCADILAA 125
+ GCD SVL+ S K E++A N + S GFEV+ K+ LE CP VSCADIL
Sbjct: 354 FVNGCDGSVLLDDTSTQKGEKNALPNKN-SIRGFEVIDTIKSALEKACPSTVSCADILTL 530
Query: 126 AARDLVVSVGGPFYELDLGRRDSLESKSIDAENKYPLPTMTNSQVIDIFTSKGFTVQEMV 185
AAR+ V GPF+ + LGRRD + +A N P P + F SKG +++
Sbjct: 531 AAREAVYLSRGPFWSVPLGRRDGTTASESEA-NNLPSPFEPLENITAKFISKGLEKKDVA 707
Query: 186 ALAGAHTIGFSHC 198
L+GAHT GF+ C
Sbjct: 708 VLSGAHTFGFAQC 746
>BP051104
Length = 566
Score = 127 bits (320), Expect = 2e-30
Identities = 73/171 (42%), Positives = 98/171 (56%)
Frame = +2
Query: 66 MIGGCDASVLVSSNSFNKAERDADINLSLSGDGFEVVTRAKNMLELECPGVVSCADILAA 125
++ GCDAS+L S E+ A N + S GF V+ K LE +CPGVVSCAD+LA
Sbjct: 68 LVNGCDASILXDDTSNFIGEQTAAAN-NRSARGFNVIDGIKANLEKQCPGVVSCADVLAL 244
Query: 126 AARDLVVSVGGPFYELDLGRRDSLESKSIDAENKYPLPTMTNSQVIDIFTSKGFTVQEMV 185
AARD VV +GGP +E+ LGRRDS + A N P P ++ S +I F ++G +V ++V
Sbjct: 245 AARDSVVQLGGPSWEVGLGRRDSTTASRGTANNTIPGPFLSLSGLITNFANQGLSVTDLV 424
Query: 186 ALAGAHTIGFSHCKQFSNRLFNFSKTTETDPKYNPEYAAGLKKLCQNYQKD 236
AL+GAHTIG + CK F ++N D + YA LK C D
Sbjct: 425 ALSGAHTIGLAQCKNFRAHIYN-------DSNIDASYAKFLKX*CPRSGND 556
>TC9166 similar to UP|Q9XFL4 (Q9XFL4) Peroxidase 3, partial (49%)
Length = 559
Score = 119 bits (298), Expect = 7e-28
Identities = 69/164 (42%), Positives = 96/164 (58%), Gaps = 1/164 (0%)
Frame = +3
Query: 6 YIFSLLCLALFPITQS-KLIPNYYQKSCPKFEDIVKQTVTDKQKTTPSTAGAALRLFFSD 64
++ SL L L + S +L N+Y K CP + VK V G+ LRLFF D
Sbjct: 57 FVLSLFMLFLIGSSNSAQLSENFYVKKCPSVFNAVKSVVHSAVAKEARMGGSLLRLFFHD 236
Query: 65 CMIGGCDASVLVSSNSFNKAERDADINLSLSGDGFEVVTRAKNMLELECPGVVSCADILA 124
C + GCD SVL+ S K E+ A N S S GF+V+ K+ +E CPGVVSCAD++A
Sbjct: 237 CFVNGCDGSVLLDDTSSFKGEKTAPPN-SNSLRGFDVIDAIKSKVEAVCPGVVSCADVVA 413
Query: 125 AAARDLVVSVGGPFYELDLGRRDSLESKSIDAENKYPLPTMTNS 168
AARD V +GGP++++ LGRRDS ++ S +A N +P+ +S
Sbjct: 414 IAARDSVAILGGPYWKVKLGRRDS-KTASFNAANSGVIPSPFSS 542
>TC14619 similar to GB|CAA72487.1|1890317|ATP26A peroxidase ATP26a
{Arabidopsis thaliana;} , partial (35%)
Length = 643
Score = 115 bits (288), Expect = 1e-26
Identities = 54/99 (54%), Positives = 72/99 (72%)
Frame = +3
Query: 226 LKKLCQNYQKDTSMSAFNDVMTPSKFDNMYFKNLKRGMGLLATDSLMGEDKRTKPFVDMY 285
L+K C NYQ + ++S FND+MTP+KFDN+YF+NL +G+GLL +D + D TKPFV +
Sbjct: 3 LQKACANYQTNPTLSVFNDIMTPNKFDNVYFQNLPKGLGLLKSDQGLFSDAVTKPFVAKF 182
Query: 286 AENQTKFFEDFGNAMRKLSVLHVKEGKDGEIRNRCDTFN 324
A +Q +FF F AM+KLS+L VK G+ GEIR RCD N
Sbjct: 183 AADQNEFFRVFTRAMQKLSLLGVKTGRRGEIRRRCDQVN 299
>AV426241
Length = 429
Score = 114 bits (284), Expect = 3e-26
Identities = 67/145 (46%), Positives = 83/145 (57%), Gaps = 1/145 (0%)
Frame = +3
Query: 3 YKSYIFS-LLCLALFPITQSKLIPNYYQKSCPKFEDIVKQTVTDKQKTTPSTAGAALRLF 61
+KS++ L+ L L +Y KSCP E IVK TV KT A LRL
Sbjct: 9 HKSFLLVFLIVLTLQAFAVHGTSVGFYSKSCPSIESIVKSTVASHVKTDFEYAAGLLRLH 188
Query: 62 FSDCMIGGCDASVLVSSNSFNKAERDADINLSLSGDGFEVVTRAKNMLELECPGVVSCAD 121
F DC + GCDAS+L++ N E+ A N SL G +EV+ AK LE +CPGVVSCAD
Sbjct: 189 FRDCFVRGCDASILIAGNG---TEKQAPPNRSLKG--YEVIDEAKAKLEAQCPGVVSCAD 353
Query: 122 ILAAAARDLVVSVGGPFYELDLGRR 146
ILA AARD VV GG +++ GRR
Sbjct: 354 ILALAARDSVVLSGGLSWQVPTGRR 428
>TC10510 similar to PIR|B56555|B56555 peroxidase , anionic, precursor - wood
tobacco {Nicotiana sylvestris;} , partial (53%)
Length = 764
Score = 107 bits (266), Expect = 4e-24
Identities = 66/194 (34%), Positives = 98/194 (50%)
Frame = +3
Query: 136 GPFYELDLGRRDSLESKSIDAENKYPLPTMTNSQVIDIFTSKGFTVQEMVALAGAHTIGF 195
GP + + LGRRDS + A + P T +I F +KG T ++MV L+GAHTIG
Sbjct: 3 GPSWTVKLGRRDSTTASRSLANTELPSFTDDLQTLISRFDNKGLTARDMVILSGAHTIGQ 182
Query: 196 SHCKQFSNRLFNFSKTTETDPKYNPEYAAGLKKLCQNYQKDTSMSAFNDVMTPSKFDNMY 255
+ C F R++N ++ D + G + + D ++A D++TP+ FDN Y
Sbjct: 183 AQCSTFRGRIYN--NASDIDAGFGSTRRRGCPSTI-SVENDKKLAAL-DLVTPNSFDNNY 350
Query: 256 FKNLKRGMGLLATDSLMGEDKRTKPFVDMYAENQTKFFEDFGNAMRKLSVLHVKEGKDGE 315
FKNL + GLL +D ++ T V Y++N T F DF AM K+ + G G
Sbjct: 351 FKNLIQKKGLLRSDQVLFRGGSTDTIVSEYSKNPTTFKSDFAAAMIKMGDIQPLTGSAGI 530
Query: 316 IRNRCDTFNNLNAN 329
IR C N L ++
Sbjct: 531 IRKICSAIN*LRSS 572
>AV414604
Length = 427
Score = 106 bits (264), Expect = 6e-24
Identities = 59/126 (46%), Positives = 75/126 (58%), Gaps = 4/126 (3%)
Frame = +2
Query: 10 LLCLALFPITQ----SKLIPNYYQKSCPKFEDIVKQTVTDKQKTTPSTAGAALRLFFSDC 65
L CLA+F L +Y+KSC + E IVK T+ + P LR+ F DC
Sbjct: 35 LACLAMFCFLGVCQGGSLRKQFYRKSCSQAEQIVKTTIQQHVSSRPELPAKLLRMHFHDC 214
Query: 66 MIGGCDASVLVSSNSFNKAERDADINLSLSGDGFEVVTRAKNMLELECPGVVSCADILAA 125
+ GCD SVL++S + N AE+DA NLSLS GF+V+ K LE +CP +VSCADILA
Sbjct: 215 FVRGCDGSVLLNSTAGNTAEKDAIPNLSLS--GFDVIDEIKEALEAKCPKIVSCADILAL 388
Query: 126 AARDLV 131
AARD V
Sbjct: 389 AARDAV 406
>TC16453 similar to UP|Q40372 (Q40372) Peroxidase precursor, partial (49%)
Length = 581
Score = 105 bits (262), Expect = 1e-23
Identities = 66/171 (38%), Positives = 90/171 (52%), Gaps = 3/171 (1%)
Frame = +2
Query: 11 LCLALFPITQSKLIPNYYQKSCPKFEDIVKQTVTDKQKTTPSTAGAALRLFFSDCMIGGC 70
L L P T + L P YY K CP+ I+K V + LRL F DC + GC
Sbjct: 71 LATFLIPSTLAGLTPGYYDKVCPQALPIIKSVVQKAINREQRIGASLLRLHFHDCFVNGC 250
Query: 71 DASVLVSSNSFNKAERDADINLSLSGDGFEVVTRAKNMLELECP-GVVSCADILAAAARD 129
D S+L+ S E+ A NL+ S GFEVV K ++ C VVSCADILA AARD
Sbjct: 251 DGSLLLDDTSSFVGEKTALPNLN-SVRGFEVVDEIKAAVDRACKRPVVSCADILAIAARD 427
Query: 130 LVVSVGGP--FYELDLGRRDSLESKSIDAENKYPLPTMTNSQVIDIFTSKG 178
V +GG +Y++ LGRRD+ + A + P P + +Q++ F ++G
Sbjct: 428 SVAILGGKQYWYQVRLGRRDARSASQAAANSNLPPPFLNFTQLLPAFQAQG 580
>AV412471
Length = 430
Score = 102 bits (254), Expect = 9e-23
Identities = 53/110 (48%), Positives = 67/110 (60%)
Frame = +2
Query: 26 NYYQKSCPKFEDIVKQTVTDKQKTTPSTAGAALRLFFSDCMIGGCDASVLVSSNSFNKAE 85
NYY SCP + IVK V+ + P+ A +R+ F DC I GCD SVL+ S N AE
Sbjct: 107 NYYLFSCPFVDPIVKNKVSTALQNDPTLAAGLIRMHFHDCFIEGCDGSVLLDSTKDNTAE 286
Query: 86 RDADINLSLSGDGFEVVTRAKNMLELECPGVVSCADILAAAARDLVVSVG 135
+D+ NLSL G+E++ K LE CPGVVSCADI+A A+RD V G
Sbjct: 287 KDSPANLSLR--GYEIIDDIKEELEKRCPGVVSCADIVAMASRDAVFFAG 430
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.319 0.134 0.387
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,840,769
Number of Sequences: 28460
Number of extensions: 55168
Number of successful extensions: 402
Number of sequences better than 10.0: 107
Number of HSP's better than 10.0 without gapping: 350
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 355
length of query: 329
length of database: 4,897,600
effective HSP length: 91
effective length of query: 238
effective length of database: 2,307,740
effective search space: 549242120
effective search space used: 549242120
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 55 (25.8 bits)
Medicago: description of AC146588.10


