

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146586.12 - phase: 0 /pseudo
(840 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
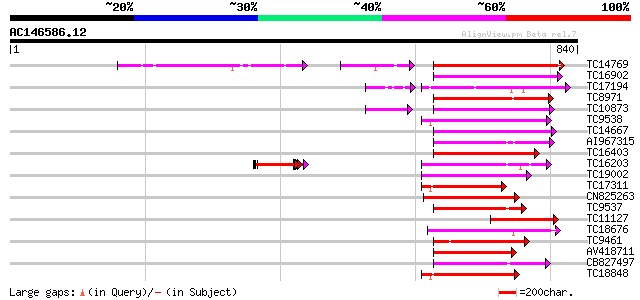
Score E
Sequences producing significant alignments: (bits) Value
TC14769 UP|Q8LKX1 (Q8LKX1) Receptor-like kinase SYMRK, complete 152 8e-38
TC16902 homologue to UP|Q8VZH5 (Q8VZH5) Receptor protein kinase-... 139 2e-33
TC17194 UP|CAE02590 (CAE02590) Nod-factor receptor 1b, complete 133 2e-33
TC8971 similar to GB|AAM16251.1|20334780|AY093990 At2g11520/F14P... 138 4e-33
TC10873 similar to UP|Q9LDZ5 (Q9LDZ5) F2D10.13 (F5M15.3), partia... 131 5e-31
TC9538 similar to UP|RLK5_ARATH (P47735) Receptor-like protein k... 131 5e-31
TC14667 homologue to UP|Q84P43 (Q84P43) Protein kinase Pti1, com... 131 5e-31
AI967315 127 1e-29
TC16403 homologue to UP|Q9LKY4 (Q9LKY4) Pti1 kinase-like protein... 124 5e-29
TC16203 UP|Q8GRU6 (Q8GRU6) LRR receptor-like kinase (Hypernodula... 123 1e-28
TC19002 weakly similar to UP|Q9XJ10 (Q9XJ10) ESTs C22458(C62866)... 123 1e-28
TC17311 similar to UP|Q8K2Y2 (Q8K2Y2) Receptor-interacting serin... 123 1e-28
CN825263 119 2e-27
TC9537 similar to UP|RLK5_ARATH (P47735) Receptor-like protein k... 118 3e-27
TC11127 weakly similar to UP|Q9FN93 (Q9FN93) Receptor-like prote... 117 1e-26
TC18676 similar to UP|Q8RYS1 (Q8RYS1) Protein kinase-like, parti... 116 2e-26
TC9461 similar to UP|O49840 (O49840) Protein kinase (AT2G02800/T... 115 2e-26
AV418711 114 8e-26
CB827497 113 1e-25
TC18848 weakly similar to UP|Q9SSQ6 (Q9SSQ6) F6D8.24 protein, pa... 110 7e-25
>TC14769 UP|Q8LKX1 (Q8LKX1) Receptor-like kinase SYMRK, complete
Length = 3495
Score = 152 bits (383), Expect(2) = 8e-38
Identities = 74/195 (37%), Positives = 121/195 (61%), Gaps = 1/195 (0%)
Frame = +3
Query: 628 MLNWNERLNIAVDAAHGLDYLHNGCKPPTMHRDLKPSNILLDENMHAKIADFGLSRAFDN 687
+L+W RL+IA+ AA GL YLH +HRD+K SNILLD +M AK+ADFG S+
Sbjct: 2445 ILDWPTRLSIALGAARGLAYLHTFPGRSVIHRDIKSSNILLDHSMCAKVADFGFSKYAPQ 2624
Query: 688 DIDSHISTRPAGTFGYVDPKFQRTGNTNKKNDIYSFGIVLLELITGKKAL-VRASGESIH 746
+ DS++S GT GY+DP++ +T ++K+D++SFG+VLLE+++G++ L ++
Sbjct: 2625 EGDSYVSLEVRGTAGYLDPEYYKTQQLSEKSDVFSFGVVLLEIVSGREPLNIKRPRTEWS 2804
Query: 747 ILQWVTPIVERGDIRSIIDARLQGKFDINSAWKVVEIAMSSTSPIEVERPDMSQILAELK 806
+++W TP + + I+D ++G + + W+VVE+A+ P RP M I+ EL+
Sbjct: 2805 LVEWATPYIRGSKVDEIVDPGIKGGYHAEAMWRVVEVALQCLEPFSTYRPSMVAIVRELE 2984
Query: 807 ECLSLDMVHRNNGRE 821
+ L ++ NN E
Sbjct: 2985 DALIIE----NNASE 3017
Score = 102 bits (253), Expect = 3e-22
Identities = 83/291 (28%), Positives = 145/291 (49%), Gaps = 10/291 (3%)
Frame = +3
Query: 160 LQYKDDVYDRMWFSYELIDWR-RLSTSLNNDHLVQNIYKPPTIVMSTAATPVNASAPLQF 218
+++ D DR+W + + LS++++N L N+ PP V+ TA T L+F
Sbjct: 984 IRFPVDQSDRIWKASSISSSAVPLSSNVSNVDLNANV-TPPLTVLQTALTDPER---LEF 1151
Query: 219 -HWSSNNENDQYYLYIHFNEVEELAANETREFNITVNDKLWFGPVTPIYRTPDLIFSTEP 277
H E+ Y ++++F E++ R F+I VN ++ + + +
Sbjct: 1152 IHTDLETEDYGYRVFLYFLELDRTLQAGQRVFDIYVNSEIKKESFDVLAGGSNYRYDVLD 1331
Query: 278 LRRAETYQISLSKTKNSTLPPILNAFEIYMAKDFSQLETQQDDVDNITNIK-------NA 330
+ + + ++L K S P+LNA+EI + + + ET Q DV I ++ +
Sbjct: 1332 ISASGSLNVTLVKASKSEFGPLLNAYEILQVRPWIE-ETNQTDVGVIQKMREELLLQNSG 1508
Query: 331 YGVTRNWQGDPCAPVNYMWEGLNCSTDDDNNPPRITSLNLSSSGLTGEISSSISKLTMLQ 390
+W GDPC + W+G+ C D N IT L+LSSS L G I SSI+++T L+
Sbjct: 1509 NRALESWSGDPC--ILLPWKGIAC--DGSNGSSVITKLDLSSSNLKGLIPSSIAEMTNLE 1676
Query: 391 YLDLSNNSLNGPLPDFLIQLRSLQV-LNVGKNNLTGLVPSELLERSKTGSL 440
L++S+NS +G +P F L SL + +++ N+L G +P +++ SL
Sbjct: 1677 TLNISHNSFDGSVPSF--PLSSLLISVDLSYNDLMGKLPESIVKLPHLKSL 1823
Score = 23.1 bits (48), Expect(2) = 8e-38
Identities = 34/113 (30%), Positives = 56/113 (49%), Gaps = 4/113 (3%)
Frame = +2
Query: 491 YNYVFKL*E*GFNKIK-TSKIQLY*NCQYY**LQNYYWRRRIWKSLLWHST---RSN*SR 546
YN++F * +K++ S I L + +QN RR+W L +S RS
Sbjct: 2072 YNFLFAKQR*FLHKVRINSSIHLGVYRGGHREVQNLDR*RRVWLCLQGYSK*WPRSGGES 2251
Query: 547 C*DAFTIINARL*GI*SRGTTSNSCSS*KFGLPCWIL**R*NQSINL*IHGQW 599
N+R+* *++ T SN+ +*+ G W+L**+*+ + ++ H QW
Sbjct: 2252 AVIDINSGNSRI**-*AKPTFSNT--A*EPGATSWLL**K*STNSSVSFHVQW 2401
>TC16902 homologue to UP|Q8VZH5 (Q8VZH5) Receptor protein kinase-like
protein, partial (46%)
Length = 941
Score = 139 bits (350), Expect = 2e-33
Identities = 74/192 (38%), Positives = 108/192 (55%), Gaps = 1/192 (0%)
Frame = +3
Query: 629 LNWNERLNIAVDAAHGLDYLHNGCKPPTMHRDLKPSNILLDENMHAKIADFGLSRAFDND 688
L W +RL I + AA GL YLH G K +HRD+K +NILLDE AK++DFGLS+
Sbjct: 276 LPWKQRLEICIGAARGLHYLHTGAKYTIIHRDVKTTNILLDEKWVAKVSDFGLSKTGPTL 455
Query: 689 IDSHISTRPAGTFGYVDPKFQRTGNTNKKNDIYSFGIVLLELITGKKAL-VRASGESIHI 747
++H+ST G+FGY+DP++ R K+D+YSFG+VL E++ + AL + E + +
Sbjct: 456 DNTHVSTVVKGSFGYLDPEYFRRQQLTDKSDVYSFGVVLFEILCARPALNPSLAKEQVSL 635
Query: 748 LQWVTPIVERGDIRSIIDARLQGKFDINSAWKVVEIAMSSTSPIEVERPDMSQILAELKE 807
+W + +G + I+D L+GK K E AM S +ERP M +L L+
Sbjct: 636 AEWASHCYNKGILDQILDPYLKGKIAPECFKKFAETAMKCVSDQGIERPSMGDVLWNLEF 815
Query: 808 CLSLDMVHRNNG 819
L L +G
Sbjct: 816 ALQLQESAEESG 851
>TC17194 UP|CAE02590 (CAE02590) Nod-factor receptor 1b, complete
Length = 2193
Score = 133 bits (334), Expect(2) = 2e-33
Identities = 75/230 (32%), Positives = 127/230 (54%), Gaps = 9/230 (3%)
Frame = +1
Query: 610 HFQNFETNAYLHAVENSNMLNWNERLNIAVDAAHGLDYLHNGCKPPTMHRDLKPSNILLD 669
H N YLH L W+ R+ IA+DAA GL+Y+H P +HRD+K +NIL+D
Sbjct: 1297 HIDNGNLGQYLHG-SGKEPLPWSSRVQIALDAARGLEYIHEHTVPVYIHRDVKSANILID 1473
Query: 670 ENMHAKIADFGLSRAFDNDIDSHISTRPAGTFGYVDPKFQRTGNTNKKNDIYSFGIVLLE 729
+N+ K+ADFGL++ + +S + TR GTFGY+ P++ + G+ + K D+Y+FG+VL E
Sbjct: 1474 KNLRGKVADFGLTKLIEVG-NSTLQTRLVGTFGYMPPEYAQYGDISPKIDVYAFGVVLFE 1650
Query: 730 LITGKKALVRAS---GESIHILQWVTPIVERGD----IRSIIDARLQGKFDINSAWKVVE 782
LI+ K A+++ ES ++ + + D +R ++D RL + I+S K+ +
Sbjct: 1651 LISAKNAVLKTGELVAESKGLVALFEEALNKSDPCDALRKLVDPRLGENYPIDSVLKIAQ 1830
Query: 783 IAMSSTSPIEVERPDMSQILAELKECLSL--DMVHRNNGRERAIVELTSL 830
+ + T + RP M ++ L SL D ++ + ++ L S+
Sbjct: 1831 LGRACTRDNPLLRPSMRSLVVALMTLSSLTEDCDDESSYESQTLINLLSV 1980
Score = 26.9 bits (58), Expect(2) = 2e-33
Identities = 30/75 (40%), Positives = 34/75 (45%), Gaps = 1/75 (1%)
Frame = +3
Query: 527 WRRRIWKSLLWHSTRSN*SRC*DAFTIINARL*GI*SRGTTSNSCSS*KFGLPCWIL**R 586
W R IW LL R S D T IN +* G N+CS + G WIL R
Sbjct: 1107 WSRWIWSCLLCRIERQENSN*EDGCTSINRI--SL*VEGL--NTCSPLESGALDWIL--R 1268
Query: 587 *NQSIN-L*IHGQWK 600
* SI L* + QWK
Sbjct: 1269 *GISIPCL*TY*QWK 1313
>TC8971 similar to GB|AAM16251.1|20334780|AY093990 At2g11520/F14P14.15
{Arabidopsis thaliana;}, partial (37%)
Length = 1079
Score = 138 bits (347), Expect = 4e-33
Identities = 70/181 (38%), Positives = 115/181 (62%), Gaps = 3/181 (1%)
Frame = +3
Query: 628 MLNWNERLNIAVDAAHGLDYLHNGCKPPTMHRDLKPSNILLDENMHAKIADFGLSRAFD- 686
+L++N+RL IA+D AHGL YLH + +HRD+K SNILL E+M AK+ADFG +R
Sbjct: 150 ILDFNQRLEIAIDVAHGLTYLHLYAEKQIIHRDVKSSNILLTESMRAKVADFGFARLGPV 329
Query: 687 NDIDSHISTRPAGTFGYVDPKFQRTGNTNKKNDIYSFGIVLLELITGKK--ALVRASGES 744
N +HIST+ GT GY+DP++ +T K+D+YSFGI+LLE++TG++ L +A+ E
Sbjct: 330 NGDQTHISTKVKGTVGYLDPEYMKTHQLTPKSDVYSFGILLLEILTGRRPVELKKAADER 509
Query: 745 IHILQWVTPIVERGDIRSIIDARLQGKFDINSAWKVVEIAMSSTSPIEVERPDMSQILAE 804
+ L+W G + ++D ++ + + K+++++ +PI +RP+M + +
Sbjct: 510 V-TLRWAFRKYNEGSVVELMDPLMEEAVNADVLMKMLDLSFQCAAPIRTDRPNMKSVGEQ 686
Query: 805 L 805
L
Sbjct: 687 L 689
>TC10873 similar to UP|Q9LDZ5 (Q9LDZ5) F2D10.13 (F5M15.3), partial (77%)
Length = 1478
Score = 131 bits (329), Expect(2) = 5e-31
Identities = 70/181 (38%), Positives = 108/181 (58%), Gaps = 3/181 (1%)
Frame = +1
Query: 629 LNWNERLNIAVDAAHGLDYLHNGCKPPTMHRDLKPSNILLDENMHAKIADFGLSRAFDND 688
LNW+ R+ +AV AA GL+YLH PP ++RDLK +NILLD + K++DFGL++
Sbjct: 736 LNWSTRMKVAVGAARGLEYLHCTADPPVIYRDLKSANILLDNEFNPKLSDFGLAKLGPVG 915
Query: 689 IDSHISTRPAGTFGYVDPKFQRTGNTNKKNDIYSFGIVLLELITGKKAL--VRASGESIH 746
++H+STR GT+GY P++ +G K+DIYSFG+VLLEL+TG++A+ R GE +
Sbjct: 916 DNTHVSTRVMGTYGYCAPEYAMSGKLTLKSDIYSFGVVLLELLTGRRAIDTSRRPGEQ-N 1092
Query: 747 ILQWVTP-IVERGDIRSIIDARLQGKFDINSAWKVVEIAMSSTSPIEVERPDMSQILAEL 805
++ W P +R ++D LQG+F + + I RP ++ I+ L
Sbjct: 1093LVSWARPYFSDRRRFGHMVDPLLQGRFPSRCLHQAIAITAMCLQEQPKFRPLITDIVVAL 1272
Query: 806 K 806
+
Sbjct: 1273E 1275
Score = 21.2 bits (43), Expect(2) = 5e-31
Identities = 19/70 (27%), Positives = 31/70 (44%)
Frame = +3
Query: 527 WRRRIWKSLLWHSTRSN*SRC*DAFTIINARL*GI*SRGTTSNSCSS*KFGLPCWIL**R 586
WRR WKSL S C + ++R G+ G+ + + W+L *
Sbjct: 471 WRRWFWKSLQGSSHDWRGCCCKTIKS*WSSRFSGVCDGGSYVKPAAPYQSC*VNWLLH*W 650
Query: 587 *NQSINL*IH 596
*+++ L*+H
Sbjct: 651 *SEAFGL*VH 680
>TC9538 similar to UP|RLK5_ARATH (P47735) Receptor-like protein kinase 5
precursor , partial (10%)
Length = 1067
Score = 131 bits (329), Expect = 5e-31
Identities = 70/203 (34%), Positives = 114/203 (55%), Gaps = 10/203 (4%)
Frame = +2
Query: 610 HFQNFETNAYLH----------AVENSNMLNWNERLNIAVDAAHGLDYLHNGCKPPTMHR 659
+ +N + +LH V+ +L+W +RL IA+ AA GL Y+H+ C PP +HR
Sbjct: 65 YLENHSLDKWLHLKPKSSSVSGVVQQYTVLDWPKRLKIAIGAAQGLSYMHHDCSPPIVHR 244
Query: 660 DLKPSNILLDENMHAKIADFGLSRAFDNDIDSHISTRPAGTFGYVDPKFQRTGNTNKKND 719
D+K SNILLD+ +AK+ADFGL+R + +I + GTFGY+ P++ +T ++K D
Sbjct: 245 DVKTSNILLDKQFNAKVADFGLARMLIKPGELNIMSTVIGTFGYIAPEYVQTTRISEKVD 424
Query: 720 IYSFGIVLLELITGKKALVRASGESIHILQWVTPIVERGDIRSIIDARLQGKFDINSAWK 779
+YSFG+VLLEL TGK+A S+ W I+ ++ ++ + I+
Sbjct: 425 VYSFGVVLLELTTGKEANYGDQHSSLAEWAW-RHILIGSNVXDLLXKDVMEASYIDEMCS 601
Query: 780 VVEIAMSSTSPIEVERPDMSQIL 802
V ++ + T+ + RP M ++L
Sbjct: 602 VFKLGVMCTATLPATRPSMKEVL 670
>TC14667 homologue to UP|Q84P43 (Q84P43) Protein kinase Pti1, complete
Length = 1591
Score = 131 bits (329), Expect = 5e-31
Identities = 70/183 (38%), Positives = 108/183 (58%), Gaps = 2/183 (1%)
Frame = +2
Query: 629 LNWNERLNIAVDAAHGLDYLHNGCKPPTMHRDLKPSNILLDENMHAKIADFGLSRAFDND 688
L+W +R+ IAVDAA GL+YLH +P +HRD++ SN+L+ E+ AKIADF LS +
Sbjct: 662 LDWIQRVRIAVDAARGLEYLHEKVQPAIIHRDIRSSNVLIFEDYKAKIADFNLSNQAPDM 841
Query: 689 IDSHISTRPAGTFGYVDPKFQRTGNTNKKNDIYSFGIVLLELITGKKAL--VRASGESIH 746
STR GTFGY P++ TG +K+D+YSFG+VLLEL+TG+K + G+
Sbjct: 842 AARLHSTRVLGTFGYHAPEYAMTGQLTQKSDVYSFGVVLLELLTGRKPVDHTMPRGQQ-S 1018
Query: 747 ILQWVTPIVERGDIRSIIDARLQGKFDINSAWKVVEIAMSSTSPIEVERPDMSQILAELK 806
++ W TP + ++ +D +L+G++ K+ +A RP+MS ++ L+
Sbjct: 1019LVTWATPRLSEDKVKQCVDPKLKGEYPPKGVAKLAAVAALCVQYEAEFRPNMSIVVKALQ 1198
Query: 807 ECL 809
L
Sbjct: 1199PLL 1207
>AI967315
Length = 1308
Score = 127 bits (318), Expect = 1e-29
Identities = 65/179 (36%), Positives = 104/179 (57%)
Frame = +1
Query: 629 LNWNERLNIAVDAAHGLDYLHNGCKPPTMHRDLKPSNILLDENMHAKIADFGLSRAFDND 688
L+W R I + A GL YLH GC+ +HRD+K SNILL E+ +I+DFGL++ +
Sbjct: 430 LDWKTRYKIVLGTARGLHYLHKGCQRRIIHRDIKASNILLTEDFEPQISDFGLAKWLPSQ 609
Query: 689 IDSHISTRPAGTFGYVDPKFQRTGNTNKKNDIYSFGIVLLELITGKKALVRASGESIHIL 748
H GTFG++ P++ G ++K D+++FG+ LLE+I+G+K V S +S+H
Sbjct: 610 WTHHSIAPIEGTFGHLAPEYYMHGVVDEKTDVFAFGVFLLEVISGRKP-VDGSHQSLH-- 780
Query: 749 QWVTPIVERGDIRSIIDARLQGKFDINSAWKVVEIAMSSTSPIEVERPDMSQILAELKE 807
W PI+ + +I ++D RL+G +D+ +V A RP MS++L ++E
Sbjct: 781 TWAKPILSKWEIEKLVDPRLEGCYDVTQFNRVAFAASLCIRASSTWRPTMSEVLEVMEE 957
>TC16403 homologue to UP|Q9LKY4 (Q9LKY4) Pti1 kinase-like protein, partial
(55%)
Length = 876
Score = 124 bits (312), Expect = 5e-29
Identities = 66/159 (41%), Positives = 98/159 (61%), Gaps = 2/159 (1%)
Frame = +3
Query: 628 MLNWNERLNIAVDAAHGLDYLHNGCKPPTMHRDLKPSNILLDENMHAKIADFGLSRAFDN 687
+L+W +R+ IAV AA GL+YLH + +HR +K SNILL ++ AKIADF LS +
Sbjct: 33 VLSWTQRVKIAVGAARGLEYLHEKAETHIIHRYIKSSNILLFDDDVAKIADFDLSNQAPD 212
Query: 688 DIDSHISTRPAGTFGYVDPKFQRTGNTNKKNDIYSFGIVLLELITGKKAL--VRASGESI 745
STR GTFGY P++ TG K+D+YSFG+VLLEL+TG+K + G+
Sbjct: 213 AAARLHSTRVLGTFGYHAPEYAMTGQLTSKSDVYSFGVVLLELLTGRKPVDHTLPRGQQ- 389
Query: 746 HILQWVTPIVERGDIRSIIDARLQGKFDINSAWKVVEIA 784
++ W TP + ++ +DARL+G++ + S K+ +A
Sbjct: 390 SLVTWATPKLSEDKVKQCVDARLKGEYPVKSVAKMAAVA 506
>TC16203 UP|Q8GRU6 (Q8GRU6) LRR receptor-like kinase (Hypernodulation aberrant
root formation protein), complete
Length = 3308
Score = 123 bits (309), Expect = 1e-28
Identities = 69/200 (34%), Positives = 112/200 (55%), Gaps = 7/200 (3%)
Frame = +2
Query: 610 HFQNFETNAYLHAVENSNMLNWNERLNIAVDAAHGLDYLHNGCKPPTMHRDLKPSNILLD 669
+ N +LH + + L W R IAV+AA GL Y+H+ C P +HRD+K +NILLD
Sbjct: 2420 YMPNGSLGEWLHGAKGGH-LRWEMRYKIAVEAARGLCYMHHDCSPLIIHRDVKSNNILLD 2596
Query: 670 ENMHAKIADFGLSRAFDNDIDSHISTRPAGTFGYVDPKFQRTGNTNKKNDIYSFGIVLLE 729
+ A +ADFGL++ + S + AG++GY+ P++ T ++K+D+YSFG+VLLE
Sbjct: 2597 ADFEAHVADFGLAKFLYDPGASQSMSSIAGSYGYIAPEYAYTLKVDEKSDVYSFGVVLLE 2776
Query: 730 LITGKKALVRASGESIHILQWVTPIV-------ERGDIRSIIDARLQGKFDINSAWKVVE 782
LI G+K V G+ + I+ WV + + + +++D RL G + + S +
Sbjct: 2777 LIIGRKP-VGEFGDGVDIVGWVNKTMSELSQPSDTALVLAVVDPRLSG-YPLTSVIHMFN 2950
Query: 783 IAMSSTSPIEVERPDMSQIL 802
IAM + RP M +++
Sbjct: 2951 IAMMCVKEMGPARPTMREVV 3010
Score = 56.2 bits (134), Expect = 2e-08
Identities = 28/80 (35%), Positives = 47/80 (58%)
Frame = +2
Query: 363 PRITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGKNN 422
P +T +N+S + LTG I ++I+ L +DLS N+L G +P + L L +LN+ +N
Sbjct: 1628 PMLTKVNISGNNLTGPIPTTITHRASLTAVDLSRNNLAGEVPKGMKNLMDLSILNLSRNE 1807
Query: 423 LTGLVPSELLERSKTGSLSL 442
++G VP E+ + +L L
Sbjct: 1808 ISGPVPDEIRFMTSLTTLDL 1867
Score = 54.3 bits (129), Expect = 8e-08
Identities = 25/65 (38%), Positives = 41/65 (62%)
Frame = +2
Query: 365 ITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGKNNLT 424
+T+++LS + L GE+ + L L L+LS N ++GP+PD + + SL L++ NN T
Sbjct: 1706 LTAVDLSRNNLAGEVPKGMKNLMDLSILNLSRNEISGPVPDEIRFMTSLTTLDLSSNNFT 1885
Query: 425 GLVPS 429
G VP+
Sbjct: 1886 GTVPT 1900
Score = 50.8 bits (120), Expect = 9e-07
Identities = 27/67 (40%), Positives = 39/67 (57%)
Frame = +2
Query: 365 ITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGKNNLT 424
+ SL+LS + LTGEI S SKL L ++ N G LP F+ L +L+ L V +NN +
Sbjct: 989 LMSLDLSINDLTGEIPESFSKLKNLTLMNFFQNKFRGSLPSFIGDLPNLETLQVWENNFS 1168
Query: 425 GLVPSEL 431
++P L
Sbjct: 1169 FVLPHNL 1189
Score = 48.5 bits (114), Expect = 4e-06
Identities = 22/64 (34%), Positives = 39/64 (60%)
Frame = +2
Query: 365 ITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGKNNLT 424
+ SL+L ++ GEI + ++ ML +++S N+L GP+P + SL +++ +NNL
Sbjct: 1562 LQSLSLDANEFIGEIPGGVFEIPMLTKVNISGNNLTGPIPTTITHRASLTAVDLSRNNLA 1741
Query: 425 GLVP 428
G VP
Sbjct: 1742 GEVP 1753
Score = 46.6 bits (109), Expect = 2e-05
Identities = 24/69 (34%), Positives = 39/69 (55%)
Frame = +2
Query: 361 NPPRITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGK 420
N ++ SL + + LTG I +S + L LDLS N L G +P+ +L++L ++N +
Sbjct: 905 NLTKLHSLFVQMNNLTGTIPPELSSMMSLMSLDLSINDLTGEIPESFSKLKNLTLMNFFQ 1084
Query: 421 NNLTGLVPS 429
N G +PS
Sbjct: 1085NKFRGSLPS 1111
Score = 44.3 bits (103), Expect = 8e-05
Identities = 22/67 (32%), Positives = 41/67 (60%), Gaps = 1/67 (1%)
Frame = +2
Query: 368 LNLSSSGLTGEISSSISK-LTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGKNNLTGL 426
LN+S + +G+ +I+ +T L+ LD +NS +GPLP+ +++L L+ L++ N +G
Sbjct: 488 LNISHNLFSGQFPGNITVGMTELEALDAYDNSFSGPLPEEIVKLEKLKYLHLAGNYFSGT 667
Query: 427 VPSELLE 433
+P E
Sbjct: 668 IPESYSE 688
Score = 40.4 bits (93), Expect = 0.001
Identities = 22/79 (27%), Positives = 42/79 (52%)
Frame = +2
Query: 364 RITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGKNNL 423
R+ + ++ + G I I + L + ++NN L+GP+P + QL S+ + + N L
Sbjct: 1274 RLKTFIITDNFFRGPIPKGIGECRSLTKIRVANNFLDGPVPPGVFQLPSVTITELSNNRL 1453
Query: 424 TGLVPSELLERSKTGSLSL 442
G +PS ++ G+L+L
Sbjct: 1454 NGELPS-VISGESLGTLTL 1507
Score = 37.4 bits (85), Expect = 0.010
Identities = 38/148 (25%), Positives = 65/148 (43%), Gaps = 14/148 (9%)
Frame = +2
Query: 295 TLPPILNAFEIYMAKDFSQLETQQDDVDNITNIKNAY----------GVTRNWQGD-PCA 343
T+PP L++ M+ D S + + ++ + +KN G ++ GD P
Sbjct: 956 TIPPELSSMMSLMSLDLSINDLTGEIPESFSKLKNLTLMNFFQNKFRGSLPSFIGDLPNL 1135
Query: 344 PVNYMWEGLNCSTDDDNN---PPRITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLN 400
+WE N S +N R +++ + LTG I + K L+ +++N
Sbjct: 1136 ETLQVWEN-NFSFVLPHNLGGNGRFLYFDVTKNHLTGLIPPDLCKSGRLKTFIITDNFFR 1312
Query: 401 GPLPDFLIQLRSLQVLNVGKNNLTGLVP 428
GP+P + + RSL + V N L G VP
Sbjct: 1313 GPIPKGIGECRSLTKIRVANNFLDGPVP 1396
>TC19002 weakly similar to UP|Q9XJ10 (Q9XJ10) ESTs C22458(C62866), partial
(48%)
Length = 780
Score = 123 bits (308), Expect = 1e-28
Identities = 63/165 (38%), Positives = 97/165 (58%), Gaps = 2/165 (1%)
Frame = +1
Query: 610 HFQNFETNAYLHAVENSN-MLNWNERLNIAVDAAHGLDYLHNGCKPPTMHRDLKPSNILL 668
+ N ++LH +S +L+WN R+NIA+ +A G+ YLH+ P +HRD+K SN+LL
Sbjct: 289 YMPNLSLLSHLHGQHSSECLLDWNRRMNIAIGSAEGIVYLHHQATPHIIHRDIKASNVLL 468
Query: 669 DENMHAKIADFGLSRAFDNDIDSHISTRPAGTFGYVDPKFQRTGNTNKKNDIYSFGIVLL 728
D + A++ADFG ++ D +H++TR GT GY+ P++ G N+ D++SFGI+LL
Sbjct: 469 DSDFQARVADFGFAKLIP-DGATHVTTRVKGTLGYLAPEYAMLGKANECCDVFSFGILLL 645
Query: 729 ELITGKKALVRASGE-SIHILQWVTPIVERGDIRSIIDARLQGKF 772
EL +GKK L + S I W P+ D RL G++
Sbjct: 646 ELASGKKPLEKLSSTVKRSINDWALPLACAKKFTEFADPRLNGEY 780
>TC17311 similar to UP|Q8K2Y2 (Q8K2Y2) Receptor-interacting serine-threonine
kinase 3, partial (7%)
Length = 606
Score = 123 bits (308), Expect = 1e-28
Identities = 61/137 (44%), Positives = 88/137 (63%), Gaps = 10/137 (7%)
Frame = +3
Query: 610 HFQNFETNAYLH----------AVENSNMLNWNERLNIAVDAAHGLDYLHNGCKPPTMHR 659
+ +N + +LH +V+N N+++W RL+IA+ A GL Y+H+ C PP +HR
Sbjct: 96 YLENLSLDRWLHKKSKPPTVSGSVQN-NIIDWPRRLHIAIGVAQGLCYMHHDCSPPVVHR 272
Query: 660 DLKPSNILLDENMHAKIADFGLSRAFDNDIDSHISTRPAGTFGYVDPKFQRTGNTNKKND 719
DLKPSNILLD +AK+ADFGL+ + + AGTFGY+ P++ T N+K D
Sbjct: 273 DLKPSNILLDSQFNAKVADFGLAMMSVKPEELATMSAVAGTFGYIAPEYALTIRVNEKID 452
Query: 720 IYSFGIVLLELITGKKA 736
+YSFG++LLEL TGK+A
Sbjct: 453 VYSFGVILLELTTGKEA 503
>CN825263
Length = 663
Score = 119 bits (299), Expect = 2e-27
Identities = 62/146 (42%), Positives = 93/146 (63%), Gaps = 3/146 (2%)
Frame = +1
Query: 613 NFETNAYLHAVEN-SNMLNWNERLNIAVDAAHGLDYLHNGCKPPTMHRDLKPSNILLDEN 671
N ++LH + + L+WN R+ IA+ AA GL YLH P +HRD K SNILL+ +
Sbjct: 205 NGSVESHLHGADKETGPLDWNARMKIALGAARGLAYLHEDSNPCVIHRDFKSSNILLECD 384
Query: 672 MHAKIADFGLSRAFDNDIDSHISTRPAGTFGYVDPKFQRTGNTNKKNDIYSFGIVLLELI 731
K++DFGL+R ++ + HIST GTFGY+ P++ TG+ K+D+YS+G+VLLEL+
Sbjct: 385 FTPKVSDFGLARTALDEGNKHISTHVMGTFGYLAPEYAMTGHLLVKSDVYSYGVVLLELL 564
Query: 732 TGKKA--LVRASGESIHILQWVTPIV 755
TG K L + G+ +++ W PI+
Sbjct: 565 TGTKPVDLSQPPGQE-NLVTWARPIL 639
>TC9537 similar to UP|RLK5_ARATH (P47735) Receptor-like protein kinase 5
precursor , partial (10%)
Length = 480
Score = 118 bits (296), Expect = 3e-27
Identities = 60/139 (43%), Positives = 88/139 (63%), Gaps = 1/139 (0%)
Frame = +1
Query: 628 MLNWNERLNIAVDAAHGLDYLHNGCKPPTMHRDLKPSNILLDENMHAKIADFGLSRAFDN 687
+L+W +RL IA+ AHGL Y+H+ C PP +HRD+K SNILLD +AK+ADFGL+R
Sbjct: 22 VLDWPKRLRIAIGVAHGLCYMHHDCSPPIVHRDIKTSNILLDTGFNAKVADFGLARMLMK 201
Query: 688 DIDSHISTRPAGTFGYVDPKFQRTGNTNKKNDIYSFGIVLLELITGKKALVRASGESIHI 747
+ + G+FGY+ P++ +T + K D+YSFG+VLLEL TGK+A E +
Sbjct: 202 SGQFNTMSAVIGSFGYMAPEYVQTTRVSVKVDVYSFGVVLLELATGKEA--NYGDEHSSL 375
Query: 748 LQWVTPIVERG-DIRSIID 765
+W V G +I+ ++D
Sbjct: 376 AEWAWRHVHVGSNIQELVD 432
>TC11127 weakly similar to UP|Q9FN93 (Q9FN93) Receptor-like protein kinase,
partial (9%)
Length = 583
Score = 117 bits (292), Expect = 1e-26
Identities = 53/100 (53%), Positives = 78/100 (78%)
Frame = +1
Query: 713 NTNKKNDIYSFGIVLLELITGKKALVRASGESIHILQWVTPIVERGDIRSIIDARLQGKF 772
N N+K+D++SFGIVLLELITG+ A+++ + +HIL+W+TP +E GD+ I+D RLQGKF
Sbjct: 13 NLNEKSDVFSFGIVLLELITGRHAVLKGN-PCMHILEWLTPELEGGDVSRILDPRLQGKF 189
Query: 773 DINSAWKVVEIAMSSTSPIEVERPDMSQILAELKECLSLD 812
D +S WK + IAMS T+P ++RP MS +LAEL++C ++
Sbjct: 190 DASSGWKALGIAMSCTAPSSIQRPTMSVVLAELRQCFRME 309
>TC18676 similar to UP|Q8RYS1 (Q8RYS1) Protein kinase-like, partial (15%)
Length = 814
Score = 116 bits (290), Expect = 2e-26
Identities = 66/210 (31%), Positives = 113/210 (53%), Gaps = 14/210 (6%)
Frame = +3
Query: 620 LHAVENS--NMLNWNERLNIAVDAAHGLDYLHNGCKPPTMHRDLKPSNILLDENMHAKIA 677
LH +N + L+W R+ IA+DAA GL+Y+H K +H+D+ SNILLD + AKI+
Sbjct: 3 LHDPQNKGHSSLSWITRVQIALDAARGLEYIHEHTKTRYVHQDINTSNILLDASFRAKIS 182
Query: 678 DFGLSRAFDNDID-SHISTRPAGTFGYVDPKFQRTGNTNKKNDIYSFGIVLLELITGKKA 736
DFGL++ I+ +T+ T+GY+ P++ K+D+Y+FG+VL E+I+GKKA
Sbjct: 183 DFGLAKLVSETIEGGTTTTKGVSTYGYLAPEYLSNRIATSKSDVYAFGVVLYEIISGKKA 362
Query: 737 LVRASGES-----------IHILQWVTPIVERGDIRSIIDARLQGKFDINSAWKVVEIAM 785
+++ G + +L+ V + IR+ +D ++ + + ++ +A
Sbjct: 363 IIQTQGTQGPERRSLASIMLEVLRTVPDSLSTPSIRNHVDPIMKDLYSHDCVLQMAMLAK 542
Query: 786 SSTSPIEVERPDMSQILAELKECLSLDMVH 815
+ RPDM Q++ LSL +H
Sbjct: 543 QCVEEDPILRPDMKQVV------LSLSQIH 614
>TC9461 similar to UP|O49840 (O49840) Protein kinase (AT2G02800/T20F6.6) ,
partial (37%)
Length = 484
Score = 115 bits (289), Expect = 2e-26
Identities = 57/143 (39%), Positives = 96/143 (66%), Gaps = 2/143 (1%)
Frame = +3
Query: 629 LNWNERLNIAVDAAHGLDYLHNGCKPPTMHRDLKPSNILLDENMHAKIADFGLSRAFDND 688
L+W+ R+ +A+ AA GL +LHN K ++RD K SNILLD +AK++DFGL++A
Sbjct: 57 LSWSVRMKVAIGAARGLSFLHNA-KSQVIYRDFKASNILLDAEFNAKLSDFGLAKAGPTG 233
Query: 689 IDSHISTRPAGTFGYVDPKFQRTGNTNKKNDIYSFGIVLLELITGKKALVRA-SGESIHI 747
+H+ST+ GT GY P++ TG K+D+YSFG+VLLEL++G++A+ + +G ++
Sbjct: 234 DRTHVSTQVMGTQGYAAPEYVATGRLTAKSDVYSFGVVLLELLSGRRAVDKTIAGVDQNL 413
Query: 748 LQWVTPIV-ERGDIRSIIDARLQ 769
+ W P + ++ + I+D++L+
Sbjct: 414 VDWARPYLGDKRRLFRIMDSKLE 482
>AV418711
Length = 419
Score = 114 bits (284), Expect = 8e-26
Identities = 52/123 (42%), Positives = 81/123 (65%), Gaps = 1/123 (0%)
Frame = +3
Query: 629 LNWNERLNIAVDAAHGLDYLHNGCKPPTMHRDLKPSNILLDENMHAKIADFGLSRAFDND 688
L+W ERL+I + +A GL YLH G +HRD+K +NILLD+N+ AK+ADFGLS+
Sbjct: 42 LSWKERLDICIGSARGLHYLHTGYAKAVIHRDVKSANILLDDNLMAKVADFGLSKTGPEL 221
Query: 689 IDSHISTRPAGTFGYVDPKFQRTGNTNKKNDIYSFGIVLLELITGKKAL-VRASGESIHI 747
+H+ST G+FGY+DP++ R +K+D+YSFG+VL E++ + + E +++
Sbjct: 222 DQTHVSTAVKGSFGYLDPEYFRRQQLTEKSDVYSFGVVLFEVLCARPVIDPSLPREMVNL 401
Query: 748 LQW 750
+W
Sbjct: 402 AEW 410
>CB827497
Length = 550
Score = 113 bits (282), Expect = 1e-25
Identities = 59/172 (34%), Positives = 96/172 (55%)
Frame = +2
Query: 629 LNWNERLNIAVDAAHGLDYLHNGCKPPTMHRDLKPSNILLDENMHAKIADFGLSRAFDND 688
L W R NIA+ A GL YLH GC+ +HRD+K +NILL E+ +I DFGL++ +
Sbjct: 8 LPWCIRHNIALGTARGLLYLHEGCQRRIIHRDIKAANILLTEDYEPQICDFGLAKWLPEN 187
Query: 689 IDSHISTRPAGTFGYVDPKFQRTGNTNKKNDIYSFGIVLLELITGKKALVRASGESIHIL 748
H + GTFGY+ P++ G ++K D+++FG++LLEL++G++AL ++
Sbjct: 188 WTHHTVAKFEGTFGYLAPEYLLHGIVDEKTDVFAFGVLLLELVSGRRAL---DFSQQSLV 358
Query: 749 QWVTPIVERGDIRSIIDARLQGKFDINSAWKVVEIAMSSTSPIEVERPDMSQ 800
W P++++ +I +ID L FD ++ A + RP + Q
Sbjct: 359 LWAKPLLKKNEIMELIDPSLADGFDSRQMNLMLLAASLCIQQSSIRRPSIRQ 514
>TC18848 weakly similar to UP|Q9SSQ6 (Q9SSQ6) F6D8.24 protein, partial (64%)
Length = 969
Score = 110 bits (276), Expect = 7e-25
Identities = 57/150 (38%), Positives = 91/150 (60%), Gaps = 4/150 (2%)
Frame = +2
Query: 610 HFQNFETNAYLH---AVENSNMLNWNERLNIAVDAAHGLDYLHNGCKPPTMHRDLKPSNI 666
+ N ++LH AVE LNW +R+ IA+ +A G+ YLH+ P +HRD+K SN+
Sbjct: 527 YMPNLSLLSHLHGQFAVEVQ--LNWQKRMKIAIGSAEGILYLHHEVTPHIIHRDIKASNV 700
Query: 667 LLDENMHAKIADFGLSRAFDNDIDSHISTRPAGTFGYVDPKFQRTGNTNKKNDIYSFGIV 726
LL+ + +ADFG ++ + SH++TR GT GY+ P++ G ++ D+YSFGI+
Sbjct: 701 LLNSDFEPLVADFGFAKLIPEGV-SHMTTRVKGTLGYLAPEYAMWGKVSESCDVYSFGIL 877
Query: 727 LLELITGKKALVR-ASGESIHILQWVTPIV 755
LLEL+TG+K + + G I +W P++
Sbjct: 878 LLELVTGRKPIEKLPGGVKRTITEWAEPLI 967
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.343 0.150 0.492
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,236,938
Number of Sequences: 28460
Number of extensions: 195860
Number of successful extensions: 2278
Number of sequences better than 10.0: 300
Number of HSP's better than 10.0 without gapping: 2099
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2192
length of query: 840
length of database: 4,897,600
effective HSP length: 98
effective length of query: 742
effective length of database: 2,108,520
effective search space: 1564521840
effective search space used: 1564521840
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.6 bits)
S2: 59 (27.3 bits)
Medicago: description of AC146586.12


