

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146586.11 - phase: 0 /pseudo
(291 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
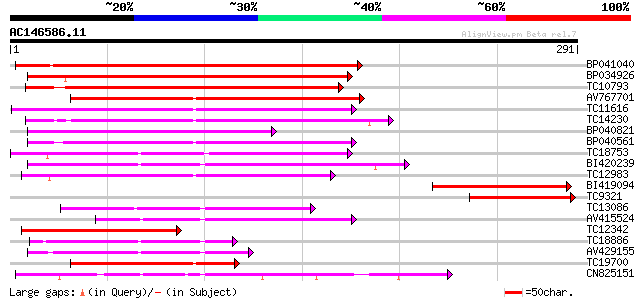
Score E
Sequences producing significant alignments: (bits) Value
BP041040 167 3e-42
BP034926 159 4e-40
TC10793 150 3e-37
AV767701 132 9e-32
TC11616 similar to UP|Q9SVU5 (Q9SVU5) Proline-rich APG-like prot... 130 2e-31
TC14230 123 3e-29
BP040821 119 5e-28
BP040561 119 5e-28
TC18753 110 3e-25
BI420239 100 5e-22
TC12983 99 8e-22
BI419094 99 1e-21
TC9321 similar to UP|O04320 (O04320) Proline-rich protein APG is... 97 4e-21
TC13086 similar to UP|Q8LDT8 (Q8LDT8) GDSL-motif lipase/hydrolas... 96 7e-21
AV415524 92 1e-19
TC12342 87 4e-18
TC18886 weakly similar to UP|Q8LDT8 (Q8LDT8) GDSL-motif lipase/h... 82 1e-16
AV429155 81 2e-16
TC19700 similar to UP|Q9SVU5 (Q9SVU5) Proline-rich APG-like prot... 80 3e-16
CN825151 79 1e-15
>BP041040
Length = 550
Score = 167 bits (422), Expect = 3e-42
Identities = 81/178 (45%), Positives = 116/178 (64%)
Frame = +3
Query: 4 MNSKETLVLLIVSCFLTCGSFAQDTLVPAIMTFGDSAVDVGNNDYLPTLFKANYPPYGRD 63
M TL L ++ FL S + VPAI+ FGDS+VD GNN+++PT+ K+N+ PYGRD
Sbjct: 3 MEKMYTLWLFLIEIFLHI-STSSSGKVPAIIVFGDSSVDSGNNNFIPTMAKSNFEPYGRD 179
Query: 64 FTNKQPTGRFCNGKLATDFTAETLGFTSFAPAYLSPQASGKNLLLGANFASAASGYDEKA 123
F + PTGRF NG++A DF +E G PAYL P S + G FASA +GYD
Sbjct: 180 FPDGNPTGRFSNGRIAPDFISEAFGLKPTIPAYLDPAYSISDFASGVCFASAGTGYDNST 359
Query: 124 ATLNHAIPLSQQLEYFKEYQGKLAQVAGSKKAASIIKDSLYVLSAGSSDFVQNYYTNP 181
+ + IPL +++EY+K+Y+ KL G +KA I+K++LY++S G++DF++NYYT P
Sbjct: 360 SNVADVIPLWKEVEYYKDYRQKLVAYLGDEKANEIVKEALYLVSIGTNDFLENYYTFP 533
>BP034926
Length = 607
Score = 159 bits (403), Expect = 4e-40
Identities = 78/177 (44%), Positives = 119/177 (67%), Gaps = 10/177 (5%)
Frame = +2
Query: 10 LVLLIVSCFLTCGSFAQD----------TLVPAIMTFGDSAVDVGNNDYLPTLFKANYPP 59
+VLL ++ + SFA D + V I+ FGDS+VD GNN+ L T K+N+PP
Sbjct: 77 VVLLALAIMMPWCSFAVDIQLARQWAAKSNVSCILVFGDSSVDPGNNNVLRTSMKSNFPP 256
Query: 60 YGRDFTNKQPTGRFCNGKLATDFTAETLGFTSFAPAYLSPQASGKNLLLGANFASAASGY 119
YG+DF N PTGRFCNG+LATDF AE LG+ PA+L P ++L G +FASAA+G+
Sbjct: 257 YGKDFFNSLPTGRFCNGRLATDFIAEALGYRQMLPAFLDPNLKVEDLPYGVSFASAATGF 436
Query: 120 DEKAATLNHAIPLSQQLEYFKEYQGKLAQVAGSKKAASIIKDSLYVLSAGSSDFVQN 176
D+ A + + +P+S+Q++YF Y+ L ++ G ++A II+++L+++S G++DF+QN
Sbjct: 437 DDYTANVVNVLPVSKQIQYFMHYKIHLRKLLGEERAEFIIRNALFIVSMGTNDFLQN 607
>TC10793
Length = 556
Score = 150 bits (378), Expect = 3e-37
Identities = 74/163 (45%), Positives = 104/163 (63%)
Frame = +1
Query: 9 TLVLLIVSCFLTCGSFAQDTLVPAIMTFGDSAVDVGNNDYLPTLFKANYPPYGRDFTNKQ 68
T +LL + C + G VPAI+ FGDS+VD GNN+++ T+ ++N+ PYGRDF +
Sbjct: 82 TTLLLQLLCLVVAGG-----KVPAIIVFGDSSVDAGNNNFIETVARSNFQPYGRDFQGGK 246
Query: 69 PTGRFCNGKLATDFTAETLGFTSFAPAYLSPQASGKNLLLGANFASAASGYDEKAATLNH 128
PTGRF NG++ATDF +E G + PAYL P + + G FASAA+GYD + +
Sbjct: 247 PTGRFSNGRIATDFISEAFGIKPYVPAYLDPSYNISHFATGVAFASAATGYDNATSDVLS 426
Query: 129 AIPLSQQLEYFKEYQGKLAQVAGSKKAASIIKDSLYVLSAGSS 171
IPL +QLEY+K YQ KL+ G KKA I SL+++S G++
Sbjct: 427 VIPLWKQLEYYKAYQKKLSTYLGEKKAHDTITKSLHIISLGTN 555
>AV767701
Length = 622
Score = 132 bits (331), Expect = 9e-32
Identities = 69/152 (45%), Positives = 94/152 (61%), Gaps = 1/152 (0%)
Frame = +3
Query: 32 AIMTFGDSAVDVGNNDYLPTLFKANYPPYGRDFTNKQPTGRFCNGKLATDFTAETLGFTS 91
A FGDS VD GNN+YL T +A+ PPYG D+ ++PTGRF NG D ++ LG S
Sbjct: 132 AFFVFGDSLVDSGNNNYLATTARADSPPYGIDYPTRRPTGRFSNGLNIPDLISQQLGAES 311
Query: 92 FAPAYLSPQASGKNLLLGANFASAASG-YDEKAATLNHAIPLSQQLEYFKEYQGKLAQVA 150
P YLSPQ G LLLGANFASA G ++ + I + +QL+YF+EYQ +LA
Sbjct: 312 VLP-YLSPQLRGNKLLLGANFASAGIGILNDTGTQFLNIIRMYRQLDYFEEYQHRLASQI 488
Query: 151 GSKKAASIIKDSLYVLSAGSSDFVQNYYTNPW 182
G K +++ +L +++ G +DFV NYY P+
Sbjct: 489 GVTKTKALVDKALVLITVGGNDFVNNYYLVPY 584
>TC11616 similar to UP|Q9SVU5 (Q9SVU5) Proline-rich APG-like protein,
partial (41%)
Length = 569
Score = 130 bits (328), Expect = 2e-31
Identities = 72/178 (40%), Positives = 103/178 (57%), Gaps = 1/178 (0%)
Frame = +2
Query: 2 MNMNSKETLVLLIVSCFLTCGSFAQDTLVPAIMTFGDSAVDVGNNDYLPTLFKANYPPYG 61
+++ S +L IV F A FGDS VD GNN+YL T +A+ PPYG
Sbjct: 38 VSLVSMASLACYIVLVVALASGFRGAEAARAFFVFGDSLVDNGNNNYLATTARADSPPYG 217
Query: 62 RDFTNKQPTGRFCNGKLATDFTAETLGFTSFAPAYLSPQASGKNLLLGANFASAASG-YD 120
D N +PTGRF NG+ DF +E LG P YLSP+ +G NLL+GANFASA G +
Sbjct: 218 IDTPNGRPTGRFSNGRNIPDFISEQLGAEPTLP-YLSPELNGDNLLVGANFASAGIGILN 394
Query: 121 EKAATLNHAIPLSQQLEYFKEYQGKLAQVAGSKKAASIIKDSLYVLSAGSSDFVQNYY 178
+ + I + +QLEYF EYQ +++ + G ++ ++ +L +++ G +DFV NYY
Sbjct: 395 DTGIQFINIIRIFRQLEYFDEYQQRVSALIGPEETQRLVNGALVLITLGGNDFVNNYY 568
>TC14230
Length = 740
Score = 123 bits (309), Expect = 3e-29
Identities = 73/192 (38%), Positives = 111/192 (57%), Gaps = 3/192 (1%)
Frame = +1
Query: 9 TLVLLIVSCFLTCGSFAQDTLVPAIMTFGDSAVDVGNNDYLPTLFKANYPPYGRDFTNKQ 68
T+ L++ FL+ S AQ T A FGDS VD GNND+L T +A+ PPYG DF +
Sbjct: 76 TITSLVLVLFLSSVS-AQHTR--AFFVFGDSLVDSGNNDFLATTARADAPPYGIDFPTHR 246
Query: 69 PTGRFCNGKLATDFTAETLGFTSFAPAYLSPQASGKNLLLGANFASAASG-YDEKAATLN 127
PTGRF NG D + LG P YLSP G+ LL+GANFASA G ++
Sbjct: 247 PTGRFSNGLNIPDLISLELGLEPTLP-YLSPLLVGEKLLIGANFASAGIGILNDTGIQFL 423
Query: 128 HAIPLSQQLEYFKEYQGKLAQVAGSKKAASIIKDSLYVLSAGSSDFVQNYYTNPWI--NQ 185
H I + +QL+ F++YQ +++ G + +++ +L++++ G +DFV NYY P+ ++
Sbjct: 424 HIIHIEKQLKLFQQYQKRVSAHIGREGTRNLVNRALFLITLGGNDFVNNYYLVPYSARSR 603
Query: 186 AITVDQYSSYLL 197
++ Y Y++
Sbjct: 604 QYSLPDYVRYII 639
>BP040821
Length = 478
Score = 119 bits (299), Expect = 5e-28
Identities = 59/128 (46%), Positives = 75/128 (58%)
Frame = +2
Query: 10 LVLLIVSCFLTCGSFAQDTLVPAIMTFGDSAVDVGNNDYLPTLFKANYPPYGRDFTNKQP 69
L+L++ S + VPA+M FGDS VD GNN+ + TL K N+PPYG+DF P
Sbjct: 92 LLLVLTSRTKAVVKLPPNVEVPAVMAFGDSIVDPGNNNKIKTLVKCNFPPYGKDFEGGNP 271
Query: 70 TGRFCNGKLATDFTAETLGFTSFAPAYLSPQASGKNLLLGANFASAASGYDEKAATLNHA 129
TGRFCNGK+ +D AE LG PAY P +LL G +FAS ASGYD +
Sbjct: 272 TGRFCNGKIPSDLLAEELGIKELLPAYKQPNLKPSDLLTGVSFASGASGYDPLTPKIASV 451
Query: 130 IPLSQQLE 137
I +S QL+
Sbjct: 452 ISMSDQLD 475
>BP040561
Length = 515
Score = 119 bits (299), Expect = 5e-28
Identities = 68/170 (40%), Positives = 96/170 (56%), Gaps = 1/170 (0%)
Frame = +1
Query: 10 LVLLIVSCFLTCGSFAQDTLVPAIMTFGDSAVDVGNNDYLPTLFKANYPPYGRDFTNKQP 69
L +++ S F+ S T A FGDS VD GNND+L T +A+ PPYG DF +P
Sbjct: 16 LCVIVTSLFMAVDS----TQTRAFFVFGDSLVDNGNNDFLATTARADAPPYGIDFPTHKP 183
Query: 70 TGRFCNGKLATDFTAETLGFTSFAPAYLSPQASGKNLLLGANFASAASG-YDEKAATLNH 128
TGRF NG D +E LG P YLSP G+ LL+GANFASA G ++ +
Sbjct: 184 TGRFSNGLNIPDLISEQLGLEPTMP-YLSPLLMGEKLLVGANFASAGIGILNDTGFQFLN 360
Query: 129 AIPLSQQLEYFKEYQGKLAQVAGSKKAASIIKDSLYVLSAGSSDFVQNYY 178
I + +QL+ F+ YQ +L+ G + ++ +L +++ G +DFV NYY
Sbjct: 361 IIHMYKQLKLFQHYQQRLSAQIGPEGTKKLVNKALVLITLGGNDFVNNYY 510
>TC18753
Length = 562
Score = 110 bits (275), Expect = 3e-25
Identities = 66/180 (36%), Positives = 101/180 (55%), Gaps = 4/180 (2%)
Frame = +3
Query: 1 MMNMNSKETLVLLIVSCF--LTCGSFAQDTLVPAIMTFGDSAVDVGNNDYLPTLFKANYP 58
+++M E +VL+++ C T + VP +FGDS VD GNN+ L +L KANY
Sbjct: 30 LVSMAHGEFIVLVLIVCLWSTTTTGVGAEPQVPCFFSFGDSLVDNGNNNRLSSLAKANYR 209
Query: 59 PYGRDFTNKQPTGRFCNGKLATDFTAETLGFTSFAPAYLSPQASGKNLLLGANFASAASG 118
PYG DF PTGRF NGK + D E LGF S+ P Y + + G+++L G N+ASAA+G
Sbjct: 210 PYGIDFPG-GPTGRFSNGKTSVDVIGELLGFGSYIPPYATTR--GQDILRGVNYASAAAG 380
Query: 119 Y-DEKAATLNHAIPLSQQLEYFKEYQGKLAQVAGSKK-AASIIKDSLYVLSAGSSDFVQN 176
+E L I Q++ ++ +L + G + AA+ + +Y + GS+D++ N
Sbjct: 381 IREETGQQLGGRISFRGQVQNYQRTVSQLINLLGDENTAANYLSKCIYSIGIGSNDYLNN 560
>BI420239
Length = 621
Score = 99.8 bits (247), Expect = 5e-22
Identities = 61/199 (30%), Positives = 108/199 (53%), Gaps = 3/199 (1%)
Frame = +1
Query: 10 LVLLIVSCFLTCGSFAQDTLVPAIMTFGDSAVDVGNNDYLPTLFKANYPPYGRDFTNKQP 69
LV+L+V+ + +++ VP + FGDS D GNN+ L T K +Y PYG DF P
Sbjct: 22 LVILLVAIAIRQYCLQRNSQVPCLFIFGDSLSDSGNNNNLETDAKVDYLPYGIDFPT-GP 198
Query: 70 TGRFCNGKLATDFTAETLGFTSFAPAYLSPQASGKNLLLGANFASAASGYDEKAAT-LNH 128
TGR+ NG+ A D E LG F P + + SG ++L G N+AS ++G +++ T L
Sbjct: 199 TGRYTNGRNAIDKITELLGLEDFIPPFAN--LSGSDILKGVNYASGSAGIRKESGTNLGT 372
Query: 129 AIPLSQQLEYFKEYQGKLAQVAGSKKAASIIKDSLYVLSAGSSDFVQNYYTNPWINQA-- 186
I + Q+ + +++ G KA + LY ++ G++D+ QNY+ + +
Sbjct: 373 NINMGLQIYFHMAIVSQISARLGFHKAKRHLSKCLYYVNIGTNDYEQNYFLPDLFDTSSK 552
Query: 187 ITVDQYSSYLLDSFTNFIK 205
T ++Y+ L++ +++++
Sbjct: 553 YTPEEYAKDLINRLSHYLQ 609
>TC12983
Length = 515
Score = 99.0 bits (245), Expect = 8e-22
Identities = 59/165 (35%), Positives = 91/165 (54%), Gaps = 4/165 (2%)
Frame = +3
Query: 7 KETLVLLIVSCFL---TCGSFAQDTLVPAIMTFGDSAVDVGNNDYLPTLFKANYPPYGRD 63
K L + I++C L T + A A GDS VD GNN+YL T +A+ PYG D
Sbjct: 24 KGCLFMSILACLLIITTWSNVAPKAEARAFFVLGDSLVDNGNNNYLATTARADAYPYGID 203
Query: 64 FTNKQPTGRFCNGKLATDFTAETLGFTSFAPAYLSPQASGKNLLLGANFASAASG-YDEK 122
+ + +GRF NG D +E +G P YLSP+ G+ LL GANFASA G ++
Sbjct: 204 SASHRASGRFSNGLNMPDLISEKIGSEPTLP-YLSPELDGEKLLNGANFASAGIGILNDT 380
Query: 123 AATLNHAIPLSQQLEYFKEYQGKLAQVAGSKKAASIIKDSLYVLS 167
+ I + QL YF++YQ +++ + G K+A +++ +L +++
Sbjct: 381 GVQFINIIRIGAQLGYFRQYQQRVSALIGEKEAQNLVNQALVLIT 515
>BI419094
Length = 585
Score = 98.6 bits (244), Expect = 1e-21
Identities = 44/71 (61%), Positives = 57/71 (79%)
Frame = +3
Query: 218 TSLPPLGCLPAARTLFGYHENGCVARINTDAQGFNKKVSSAASNLQKQLPGLKIVIFDIY 277
T+L P+GCLPAA TLFG+ N CVAR+N DA FN+K+++ + +LQK LPGLK+V+ DIY
Sbjct: 3 TTLAPVGCLPAAITLFGHDSNQCVARLNNDAVNFNRKLNTTSQSLQKSLPGLKLVLLDIY 182
Query: 278 KPLYDLVQNPS 288
+PLYDLV PS
Sbjct: 183 QPLYDLVTKPS 215
>TC9321 similar to UP|O04320 (O04320) Proline-rich protein APG isolog,
partial (37%)
Length = 541
Score = 96.7 bits (239), Expect = 4e-21
Identities = 45/54 (83%), Positives = 49/54 (90%)
Frame = +2
Query: 237 ENGCVARINTDAQGFNKKVSSAASNLQKQLPGLKIVIFDIYKPLYDLVQNPSNF 290
ENGCV+R NTDAQ FNKK++SAA+ LQKQ PGLKIVIFDIYKPLYDLVQ PSNF
Sbjct: 2 ENGCVSRFNTDAQAFNKKINSAAAKLQKQHPGLKIVIFDIYKPLYDLVQTPSNF 163
>TC13086 similar to UP|Q8LDT8 (Q8LDT8) GDSL-motif lipase/hydrolase-like
protein, partial (35%)
Length = 524
Score = 95.9 bits (237), Expect = 7e-21
Identities = 57/132 (43%), Positives = 75/132 (56%), Gaps = 1/132 (0%)
Frame = +3
Query: 27 DTLVPAIMTFGDSAVDVGNNDYLPTLFKANYPPYGRDFTNKQPTGRFCNGKLATDFTAET 86
D VP FGDS VD GNN+ L + KANY PYG DF PTGRF NGK D AE
Sbjct: 138 DPQVPCYFIFGDSLVDNGNNNQLNS*AKANYLPYGIDF-GGGPTGRFSNGKTTVDVVAEL 314
Query: 87 LGFTSFAPAYLSPQASGKNLLLGANFASAASGY-DEKAATLNHAIPLSQQLEYFKEYQGK 145
LGF S+ P Y + A G+++L G N+ASAA+G +E L I S Q+E ++ +
Sbjct: 315 LGFDSYIPPYST--ARGQDILKGVNYASAAAGIREETGQQLGGRISFSGQVENYQRTVSQ 488
Query: 146 LAQVAGSKKAAS 157
+ + G + A+
Sbjct: 489 VVNLLGDENTAA 524
>AV415524
Length = 411
Score = 91.7 bits (226), Expect = 1e-19
Identities = 52/136 (38%), Positives = 81/136 (59%), Gaps = 2/136 (1%)
Frame = +2
Query: 45 NNDYLPTLFKANYPPYGRDFTNKQPTGRFCNGKLATDFTAETLGFTSFAPAYLSPQASGK 104
+N+ L +L +A+Y PYG DF P+GRF NGK D AE LGF F P Y+S SG
Sbjct: 2 SNNALRSLARADYMPYGIDFPGG-PSGRFSNGKTTVDVIAELLGFDDFIPPYVS--TSGD 172
Query: 105 NLLLGANFASAASGY-DEKAATLNHAIPLSQQLEYFKEYQGKLAQVAGSK-KAASIIKDS 162
++L G NFASAA+G +E L I S Q++ ++ ++ + G++ +AA+ +
Sbjct: 173 DILRGVNFASAAAGIREETGQQLGGRISFSGQVQNYQSTVSQVVNILGNEDQAANYLSKC 352
Query: 163 LYVLSAGSSDFVQNYY 178
+Y + GS+D++ NY+
Sbjct: 353 IYSIGLGSNDYLNNYF 400
>TC12342
Length = 541
Score = 86.7 bits (213), Expect = 4e-18
Identities = 38/82 (46%), Positives = 54/82 (65%)
Frame = +3
Query: 7 KETLVLLIVSCFLTCGSFAQDTLVPAIMTFGDSAVDVGNNDYLPTLFKANYPPYGRDFTN 66
++TL+ + C L + VPAI+ FGDS VD GNN+++ T+ ++N+ PYGRDF
Sbjct: 288 RKTLLCFPILCILVLQLAKTNAKVPAIIVFGDSTVDAGNNNFIQTVARSNFQPYGRDFEG 467
Query: 67 KQPTGRFCNGKLATDFTAETLG 88
+ TGRFCNG++ TDF AE G
Sbjct: 468 GKATGRFCNGRIPTDFIAEDFG 533
>TC18886 weakly similar to UP|Q8LDT8 (Q8LDT8) GDSL-motif
lipase/hydrolase-like protein, partial (23%)
Length = 547
Score = 82.0 bits (201), Expect = 1e-16
Identities = 47/107 (43%), Positives = 62/107 (57%)
Frame = +1
Query: 11 VLLIVSCFLTCGSFAQDTLVPAIMTFGDSAVDVGNNDYLPTLFKANYPPYGRDFTNKQPT 70
++L+V+C + VP + FGDS D GNN+ LPTL KAN+ PYG DF PT
Sbjct: 76 LVLMVAC-MQHSVLGNSQAVPCLFVFGDSLADSGNNNNLPTLSKANFLPYGIDFPT-GPT 249
Query: 71 GRFCNGKLATDFTAETLGFTSFAPAYLSPQASGKNLLLGANFASAAS 117
GR+ NG D A+ LGF F P + + SG ++L G N+ASA S
Sbjct: 250 GRYTNGLNPIDKLAQILGFEKFIPPFAN--LSGSDILKGVNYASAFS 384
>AV429155
Length = 393
Score = 80.9 bits (198), Expect = 2e-16
Identities = 48/116 (41%), Positives = 65/116 (55%)
Frame = +3
Query: 10 LVLLIVSCFLTCGSFAQDTLVPAIMTFGDSAVDVGNNDYLPTLFKANYPPYGRDFTNKQP 69
L+LL+ +C S VP + FGDS D GNN+ L T K+NY PYG DF P
Sbjct: 54 LLLLVANCMQH--SVHGKPEVPCLFIFGDSLSDNGNNNNLVTSTKSNYKPYGIDFPT-GP 224
Query: 70 TGRFCNGKLATDFTAETLGFTSFAPAYLSPQASGKNLLLGANFASAASGYDEKAAT 125
TGRF NG D ++ LGF +F P + + S ++L G N+ASAA+G ++ T
Sbjct: 225 TGRFTNGPTTVDIISQLLGFENFIPPFAN--TSDSDILKGVNYASAAAGICVESGT 386
>TC19700 similar to UP|Q9SVU5 (Q9SVU5) Proline-rich APG-like protein,
partial (25%)
Length = 442
Score = 80.5 bits (197), Expect = 3e-16
Identities = 44/87 (50%), Positives = 53/87 (60%)
Frame = +1
Query: 32 AIMTFGDSAVDVGNNDYLPTLFKANYPPYGRDFTNKQPTGRFCNGKLATDFTAETLGFTS 91
A FGDS VD GNN+YL T +A+ PYG D+ + TGRF NG D +E +G
Sbjct: 163 AFFVFGDSLVDNGNNNYLITTARADSYPYGIDYPTHRATGRFSNGLNIPDLISERIGSEP 342
Query: 92 FAPAYLSPQASGKNLLLGANFASAASG 118
P YLSPQ G+ LL+GANFASA G
Sbjct: 343 TLP-YLSPQLDGEALLVGANFASAGIG 420
>CN825151
Length = 740
Score = 78.6 bits (192), Expect = 1e-15
Identities = 67/244 (27%), Positives = 99/244 (40%), Gaps = 20/244 (8%)
Frame = +1
Query: 4 MNSKETLVLLIVSCFLTCGSF----AQDTLVPAIMTFGDSAVDVGNNDYLPTLFKANYPP 59
M + + +SC L S + PAI FGDS D G + F+ PP
Sbjct: 7 MGLRPLFIAFFLSCTLCVNSVELKNSPPCAFPAIYNFGDSNSDTGG---ISAAFEPIPPP 177
Query: 60 YGRDFTNKQPTGRFCNGKLATDFTAETLGFTSFAPAYLSPQASGKNLLLGANFASAASGY 119
YG F K P+ R C+G+L DF AE L + AYL+ + G N GANFA+ S
Sbjct: 178 YGESFPQK-PSARDCDGRLIVDFIAEKLNLP-YLSAYLN--SLGTNYRHGANFATGGSTI 345
Query: 120 DEKAATLNH----AIPLSQQLEYFKEYQGKLAQVAGSKKAA---------SIIKDSLYVL 166
+ T+ L Q FK+++ + Q+ K A +LY
Sbjct: 346 RRQNETIFQYGISPFSLDMQFVQFKQFKARTKQLYQEAKTALERSKLPVPEEFSKALYTF 525
Query: 167 SAGSSDFVQNYYTNPWINQAITVDQYSSYLLD---SFTNFIKGVYGLGARKIGVTSLPPL 223
G +D + + + DQ + D + +K +Y LG R + + P+
Sbjct: 526 DIGQNDLSVGF-------RMMNFDQMRESMPDIVNQLASAVKNIYELGGRTFWIHNTAPI 684
Query: 224 GCLP 227
GCLP
Sbjct: 685 GCLP 696
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.318 0.135 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,379,749
Number of Sequences: 28460
Number of extensions: 50851
Number of successful extensions: 303
Number of sequences better than 10.0: 80
Number of HSP's better than 10.0 without gapping: 270
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 271
length of query: 291
length of database: 4,897,600
effective HSP length: 89
effective length of query: 202
effective length of database: 2,364,660
effective search space: 477661320
effective search space used: 477661320
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 55 (25.8 bits)
Medicago: description of AC146586.11


