

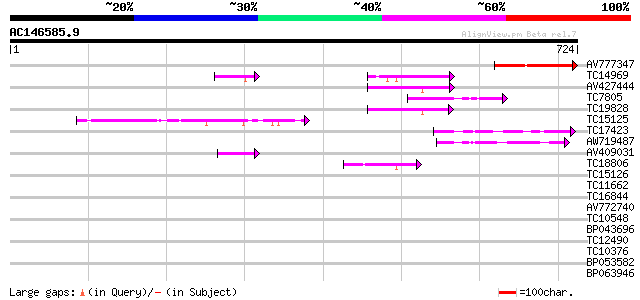
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146585.9 - phase: 0
(724 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
AV777347 169 2e-42
TC14969 similar to PIR|PQ0772|PQ0772 4-coumarate-CoA ligase (cl... 73 1e-13
AV427444 73 2e-13
TC7805 homologue to PIR|PQ0772|PQ0772 4-coumarate-CoA ligase (c... 62 2e-10
TC19828 similar to UP|Q9FNT6 (Q9FNT6) Long chain acyl-CoA synthe... 55 5e-08
TC15125 weakly similar to UP|Q84P20 (Q84P20) Acyl-activating enz... 50 1e-06
TC17423 similar to PIR|H85064|H85064 4-coumarate-CoA ligase-like... 48 5e-06
AW719487 44 7e-05
AV409031 44 1e-04
TC18806 similar to UP|Q8S564 (Q8S564) 4-coumarate:coenzyme A lig... 42 3e-04
TC15126 similar to UP|Q84P19 (Q84P19) Acyl-activating enzyme 11,... 40 0.001
TC11662 similar to UP|O65748 (O65748) Acyl-CoA synthetase (Frag... 40 0.001
TC16844 similar to UP|Q8LRT6 (Q8LRT6) Adenosine monophosphate bi... 39 0.002
AV772740 39 0.003
TC10548 similar to UP|Q940V0 (Q940V0) T23O15.3/T23O15.3, partial... 38 0.007
BP043696 38 0.007
TC12490 similar to UP|Q9FFE6 (Q9FFE6) AMP-binding protein (Adeno... 37 0.014
TC10376 similar to UP|Q8VZF1 (Q8VZF1) AT3g16910/K14A17_3 (Adenos... 35 0.032
BP053582 32 0.039
BP063946 34 0.094
>AV777347
Length = 558
Score = 169 bits (427), Expect = 2e-42
Identities = 87/105 (82%), Positives = 97/105 (91%)
Frame = -3
Query: 620 IVVIGQDKRRLGAIIVPNSEEVLKVARELSIIDSISSNVVSEEKVLNLIYKELKTWMSES 679
IVVIGQDKRRLGA+IVPN EEVLK ARELSIIDS SS+V S+EKV NLI+KELKTWMS S
Sbjct: 556 IVVIGQDKRRLGAVIVPNKEEVLKAARELSIIDSDSSDV-SQEKVTNLIHKELKTWMSGS 380
Query: 680 PFQIGPILLVNEPFTIDNGLMTPTMKIRRDRVVAKYKEQIDDLYK 724
FQIGPILLVN+PFTIDNGL+TPTMKIRRDRVVA+Y+ QI++LYK
Sbjct: 379 TFQIGPILLVNDPFTIDNGLLTPTMKIRRDRVVAQYRNQIENLYK 245
>TC14969 similar to PIR|PQ0772|PQ0772 4-coumarate-CoA ligase (clone
GM4CL1B) - soybean (fragment) {Glycine
max;} , partial (70%)
Length = 928
Score = 73.2 bits (178), Expect = 1e-13
Identities = 45/122 (36%), Positives = 67/122 (54%), Gaps = 10/122 (8%)
Frame = +2
Query: 457 ISGGGSLPLEVDKFFEAIGVKVQN-----GYGLTETSPVIA-----ARRPRCNVIGSVGH 506
+SG L E++ EA+ +V GYG+TE PV++ A++P GS G
Sbjct: 569 LSGAAPLGKELE---EALRSRVPQAVLGQGYGMTEAGPVLSMSLGFAKQPFPTKSGSCGT 739
Query: 507 PVQHTEFKVVDSETGEVLPPGSKGILKVRGPPVMNGYYKNPLATNQALDKDGWLNTGDLG 566
V++ E KV+D ETG L G + +RG +M GY + AT +D +GWL+TGD+G
Sbjct: 740 VVRNAELKVLDPETGCSLGYNQPGEICIRGQQIMKGYLNDEKATATTIDAEGWLHTGDVG 919
Query: 567 WI 568
+I
Sbjct: 920 YI 925
Score = 41.6 bits (96), Expect = 5e-04
Identities = 23/62 (37%), Positives = 34/62 (54%), Gaps = 4/62 (6%)
Frame = +2
Query: 262 INSDDIATLIYTSGTTGNPKGVMLTHRNLLHQIKNLWD----TVPAEVGDRFLSMLPPWH 317
IN +D L ++SGTTG PKGV+LTH++L + D + D L +LP +H
Sbjct: 185 INPEDAVALPFSSGTTGLPKGVVLTHKSLTTSVGQQVDGENPNLSLGTEDVLLCVLPLFH 364
Query: 318 AY 319
+
Sbjct: 365 IF 370
>AV427444
Length = 428
Score = 72.8 bits (177), Expect = 2e-13
Identities = 42/117 (35%), Positives = 64/117 (53%), Gaps = 5/117 (4%)
Frame = +1
Query: 457 ISGGGSLPLEVDKFFE-AIGVKVQNGYGLTETSPVIAARRPRCNVIGSVGHPVQHTEFKV 515
+SG L ++ +F + G +V GYG+TET+ VI+ + G VG P E K+
Sbjct: 25 VSGASPLSPDIMEFLKICFGGRVTEGYGMTETTCVISCIDQGDKLGGHVGSPSPACEVKL 204
Query: 516 VDSETGEVLP---PGSKGILKVRGPPVMNGYYKNPLATNQALDKDGWLNTGDLG-WI 568
VD P +G + VRGP + GYYK+ T + +D++GWL+TGD+G W+
Sbjct: 205 VDVPEMNYTSDDQPQPRGEICVRGPIIFKGYYKDEAQTREVIDEEGWLHTGDIGTWL 375
>TC7805 homologue to PIR|PQ0772|PQ0772 4-coumarate-CoA ligase (clone
GM4CL1B) - soybean (fragment) {Glycine
max;} , partial (41%)
Length = 965
Score = 62.4 bits (150), Expect = 2e-10
Identities = 42/127 (33%), Positives = 61/127 (47%)
Frame = +3
Query: 509 QHTEFKVVDSETGEVLPPGSKGILKVRGPPVMNGYYKNPLATNQALDKDGWLNTGDLGWI 568
++ E KV+D ETG L G + +RG +M GY + AT +D +GWL+TGD+G+I
Sbjct: 90 RNAELKVLDPETGCSLGYNQPGEICIRGQQIMKGYLNDEKATATTIDAEGWLHTGDVGYI 269
Query: 569 APHHSTGRSRNSSGVIVVDGRAKDTIVLSTGENVEPAELEEAAMRSSIIQQIVVIGQDKR 628
+ + +VD R K+ I G V PAELE + I V+ Q
Sbjct: 270 ---------DDDDEIFIVD-RVKELIKFK-GFQVPPAELEGLLVSHPSIADAAVVPQKDA 416
Query: 629 RLGAIIV 635
G + V
Sbjct: 417 AAGEVPV 437
>TC19828 similar to UP|Q9FNT6 (Q9FNT6) Long chain acyl-CoA synthetase ,
partial (17%)
Length = 378
Score = 54.7 bits (130), Expect = 5e-08
Identities = 39/118 (33%), Positives = 57/118 (48%), Gaps = 8/118 (6%)
Frame = +1
Query: 457 ISGGGSLPLEVDKFFE-AIGVKVQNGYGLTETSPVIAARRPRCNVIGSVGHPVQHTEFKV 515
+SGG L + +F +G + GYGLTET +G VG P+ + K+
Sbjct: 16 LSGGAPLSADTQRFINICLGAPIGQGYGLTETCAGGTFSDFDDTSVGRVGPPLPCSYIKL 195
Query: 516 VDSETGEVLP---PGSKGILKVRGPPVMNGYYKNPLATNQA--LDKDG--WLNTGDLG 566
+D G P +G + V GP V GY+KN TN++ +D+ G W TGD+G
Sbjct: 196 IDWPEGGYSTNDSPMPRGEIVVGGPNVTLGYFKNEEKTNESYKVDERGVRWFYTGDIG 369
>TC15125 weakly similar to UP|Q84P20 (Q84P20) Acyl-activating enzyme 12,
partial (48%)
Length = 1164
Score = 50.1 bits (118), Expect = 1e-06
Identities = 67/313 (21%), Positives = 127/313 (40%), Gaps = 16/313 (5%)
Frame = +1
Query: 86 RSSAEKYGDKVALVDQYHHPPSTITYNQLEQAILDYAEGLRVIGVRPDEKLALFADNSCR 145
+ +A Y ++V+++ H T++Q + A LR + + ++ +++ A N
Sbjct: 163 KRAAASYPNRVSVI----HEGIRFTWSQTYERCRRLAFTLRSLNIAKNDVVSVLAPNIPA 330
Query: 146 WLVADQGMMASGAINVVRGSRSSVEELLQIYNHSESVALAVDGPEMFNRIAKPFYSKTGM 205
+ +GA+ SR + + I +HSE+ VD E N+ ++ +
Sbjct: 331 MYEMHFAVPMAGAVLNTINSRLDAKNIATILSHSEAKIFFVDY-EYVNK------AREAL 489
Query: 206 RFIILLWGEKSDLNLIAEENKEVPIFSFMEVIDLGRESRMALSDS----HEASQRYVYEA 261
R +++ G K N ++ +P+ ++ I+ R+ + H + YV E
Sbjct: 490 RMLVVQKGVKP--NSTEQDYSTLPLVIVIDDINSPTGVRLGELEYEQMVHRGNPNYVPEE 663
Query: 262 INSD-DIATLIYTSGTTGNPKGVMLTHRNLLHQIKNL---WDTVPAEVGDRFLSMLPPWH 317
I + L YTSGTT PKGV+ +HR +L W+ V +L LP +H
Sbjct: 664 IQEEWSPIALNYTSGTTSEPKGVVYSHRGAYLSTLSLVLGWEMGSEPV---YLWTLPMFH 834
Query: 318 AYERACEYFIFTCGIEQ---VYTTVRN-----LKDDLGRYQPHYMISVPLVFETLYSGIQ 369
C + F G+ +RN + + + +M P+VF +
Sbjct: 835 -----CNGWTFPWGVAARGGTNVCIRNCAASDIYRSISLHNVTHMCCAPIVFNII----- 984
Query: 370 KQISTSPPVRKLV 382
+ P RK++
Sbjct: 985 --LGAKPSERKVI 1017
>TC17423 similar to PIR|H85064|H85064 4-coumarate-CoA ligase-like protein
[imported] - Arabidopsis thaliana
{Arabidopsis thaliana;}, partial (25%)
Length = 655
Score = 48.1 bits (113), Expect = 5e-06
Identities = 46/181 (25%), Positives = 79/181 (43%)
Frame = +1
Query: 542 GYYKNPLATNQALDKDGWLNTGDLGWIAPHHSTGRSRNSSGVIVVDGRAKDTIVLSTGEN 601
GY+ NP AT LDK GW++TGD+G+ + G + V R K+ ++ G
Sbjct: 16 GYHNNPEATKLTLDKKGWVHTGDVGYF----------DEDGQLYVVDRIKE-LIKYKGFQ 162
Query: 602 VEPAELEEAAMRSSIIQQIVVIGQDKRRLGAIIVPNSEEVLKVARELSIIDSISSNVVSE 661
V PAELE + I L A +VP ++ ++ S +S
Sbjct: 163 VAPAELEGLLVSHPEI------------LDAAVVPYPDDEAGEVPIAYVVRSPNS----- 291
Query: 662 EKVLNLIYKELKTWMSESPFQIGPILLVNEPFTIDNGLMTPTMKIRRDRVVAKYKEQIDD 721
+L K+++ +++ Q+ P + I + T + K+ R ++AK K +I D
Sbjct: 292 ----SLTEKQIQEFIAS---QVAPFKKLRRVTFISSVPKTASGKLLRRELIAKAKSKI*D 450
Query: 722 L 722
+
Sbjct: 451 I 453
>AW719487
Length = 498
Score = 44.3 bits (103), Expect = 7e-05
Identities = 42/169 (24%), Positives = 71/169 (41%)
Frame = +2
Query: 546 NPLATNQALDKDGWLNTGDLGWIAPHHSTGRSRNSSGVIVVDGRAKDTIVLSTGENVEPA 605
+P AT + +DK+GWL+TGD+G+I + +VD R K+ ++ G V PA
Sbjct: 5 DPEATERTIDKEGWLHTGDIGYI---------DEDDELFIVD-RLKE-LIKYKGFQVAPA 151
Query: 606 ELEEAAMRSSIIQQIVVIGQDKRRLGAIIVPNSEEVLKVARELSIIDSISSNVVSEEKVL 665
ELE + I + V VP +E ++ S +E+++
Sbjct: 152 ELEALILSHPKISDVAV------------VPMKDEAAGEVPVAFVVRSNGCTDTTEDEIK 295
Query: 666 NLIYKELKTWMSESPFQIGPILLVNEPFTIDNGLMTPTMKIRRDRVVAK 714
+ K++ + +N F ID +P+ KI R + AK
Sbjct: 296 QFVSKQVVFYKR-----------INRVFFIDAIPKSPSGKILRKDLRAK 409
>AV409031
Length = 343
Score = 43.5 bits (101), Expect = 1e-04
Identities = 20/54 (37%), Positives = 30/54 (55%)
Frame = +3
Query: 266 DIATLIYTSGTTGNPKGVMLTHRNLLHQIKNLWDTVPAEVGDRFLSMLPPWHAY 319
DIA ++TSGTT PKGV L+ NL ++N+ D + +LP +H +
Sbjct: 117 DIALFLHTSGTTSRPKGVPLSQHNLASSVRNIQSVYRLTESDSTVIVLPLFHVH 278
>TC18806 similar to UP|Q8S564 (Q8S564) 4-coumarate:coenzyme A ligase ,
partial (26%)
Length = 529
Score = 42.0 bits (97), Expect = 3e-04
Identities = 30/105 (28%), Positives = 54/105 (50%), Gaps = 6/105 (5%)
Frame = +1
Query: 427 IATILFPIHLLAIKFVYSKIHSAIGLSKAGISGGGSLPLEVDKFFEAI--GVKVQNGYGL 484
+A+ + PI L +K + + + + I+G + +E+++ +A K+ GYG+
Sbjct: 103 VASFVPPIVLALVKSGEAHQYDLSSI-RVMITGAAPMGMELEEAVKARLPQAKLGQGYGM 279
Query: 485 TETSPVIA----ARRPRCNVIGSVGHPVQHTEFKVVDSETGEVLP 525
TE P+ A+ P G+ G V++ E K+VD+ETG LP
Sbjct: 280 TEAGPLAISLAFAKEPFKTKSGACGTVVRNAEMKIVDTETGASLP 414
>TC15126 similar to UP|Q84P19 (Q84P19) Acyl-activating enzyme 11, partial
(25%)
Length = 749
Score = 40.4 bits (93), Expect = 0.001
Identities = 44/208 (21%), Positives = 90/208 (43%), Gaps = 5/208 (2%)
Frame = +2
Query: 86 RSSAEKYGDKVALVDQYHHPPSTITYNQLEQAILDYAEGLRVIGVRPDEKLALFADNSCR 145
+ +A Y ++ +++ H + T++Q A LR + + ++ +++ A N
Sbjct: 149 KRAAASYPNRTSVI----HEGTWFTWSQTYDRCRRLAFSLRSLNIARNDVVSVLAPNIPA 316
Query: 146 WLVADQGMMASGAINVVRGSRSSVEELLQIYNHSESVALAVDGPEMFNRIAKPFYSKTGM 205
+ +GA+ +R + + I HSE+ VD E N+ ++ +
Sbjct: 317 MYEMHFAVPMAGAVLNTINTRLDAKNIATILRHSEAKVFFVDY-EYVNK------AREAL 475
Query: 206 RFIILLWGEKSDLNLIAEENKEVPIFSFMEVIDLGRESRMALSDSHEASQR----YVYEA 261
+ +++ G ++ N + +P+ ++ I+ R+ + + Q YV E
Sbjct: 476 KMLVVQKG--AEPNPKEQNYATLPLVIVIDDINSPTGVRLGELEYEQMVQHGNPNYVPEE 649
Query: 262 INSD-DIATLIYTSGTTGNPKGVMLTHR 288
I + L YTSGTT PKGV+ +HR
Sbjct: 650 IQEEWSPMALNYTSGTTSEPKGVVYSHR 733
>TC11662 similar to UP|O65748 (O65748) Acyl-CoA synthetase (Fragment) ,
partial (81%)
Length = 640
Score = 40.0 bits (92), Expect = 0.001
Identities = 35/122 (28%), Positives = 58/122 (46%), Gaps = 4/122 (3%)
Frame = +1
Query: 607 LEEAAMRSSIIQQIVVIGQDKRR-LGAIIVPNSEEVLKVARELSI---IDSISSNVVSEE 662
LE + S I+ I V G L A++ P + + A E I DS+ + ++
Sbjct: 7 LENIYGQVSSIESIWVYGNSLESFLVAVVTPRQQALAHWAGENGISLDFDSLCQDSRAKS 186
Query: 663 KVLNLIYKELKTWMSESPFQIGPILLVNEPFTIDNGLMTPTMKIRRDRVVAKYKEQIDDL 722
VL + K K + I + L PF ++ L+TPT K +R +++ Y+E ID++
Sbjct: 187 YVLGELSKIAKEKKLKGFEFIRAVHLDPVPFDMERDLITPTYKKKRPQLLKYYQEIIDNM 366
Query: 723 YK 724
YK
Sbjct: 367 YK 372
>TC16844 similar to UP|Q8LRT6 (Q8LRT6) Adenosine monophosphate binding
protein 1 AMPBP1, partial (25%)
Length = 663
Score = 39.3 bits (90), Expect = 0.002
Identities = 23/70 (32%), Positives = 34/70 (47%)
Frame = +3
Query: 556 KDGWLNTGDLGWIAPHHSTGRSRNSSGVIVVDGRAKDTIVLSTGENVEPAELEEAAMRSS 615
K GW TGDLG ++ G I + R+KD I++S GEN+ ELE
Sbjct: 15 KGGWFRTGDLG----------VKHPDGYIELKDRSKD-IIISGGENISSIELEGVIFSHP 161
Query: 616 IIQQIVVIGQ 625
+ + V+G+
Sbjct: 162 AVLEAAVVGR 191
>AV772740
Length = 555
Score = 38.9 bits (89), Expect = 0.003
Identities = 15/32 (46%), Positives = 23/32 (71%)
Frame = -2
Query: 266 DIATLIYTSGTTGNPKGVMLTHRNLLHQIKNL 297
+I T++YTSGTTG P V+LTH N+ ++ +
Sbjct: 329 NICTIMYTSGTTGTPSAVVLTHANITSFVRGM 234
>TC10548 similar to UP|Q940V0 (Q940V0) T23O15.3/T23O15.3, partial (10%)
Length = 760
Score = 37.7 bits (86), Expect = 0.007
Identities = 19/59 (32%), Positives = 38/59 (64%)
Frame = +3
Query: 665 LNLIYKELKTWMSESPFQIGPILLVNEPFTIDNGLMTPTMKIRRDRVVAKYKEQIDDLY 723
++ + K K SE P +I L+ +P+T ++GL+T +K++R+++ AK+K+ + LY
Sbjct: 48 ISKVAKASKLEKSEIPKKIK---LLPDPWTPESGLVTAALKLKREQLKAKFKDDLQKLY 215
>BP043696
Length = 470
Score = 37.7 bits (86), Expect = 0.007
Identities = 15/44 (34%), Positives = 29/44 (65%)
Frame = +1
Query: 109 ITYNQLEQAILDYAEGLRVIGVRPDEKLALFADNSCRWLVADQG 152
++Y+ + +A+ +A GL+ +G +E++A+FAD W +A QG
Sbjct: 313 LSYDAVFEAVTSFASGLKELGHGREERVAIFADTREEWFIALQG 444
>TC12490 similar to UP|Q9FFE6 (Q9FFE6) AMP-binding protein (Adenosine
monophosphate binding protein 5 AMPBP5), partial (21%)
Length = 387
Score = 36.6 bits (83), Expect = 0.014
Identities = 26/119 (21%), Positives = 50/119 (41%), Gaps = 3/119 (2%)
Frame = +3
Query: 556 KDGWLNTGDLGWIAPHHSTGRSRNSSGVIVVDGRAKDTIVLSTGENVEPAELEEAAMRSS 615
K+GW TGD+G + + G + + R+KD +++S GEN+ E+E
Sbjct: 27 KNGWFYTGDVGVM----------HEDGYLEIKDRSKD-VIISGGENLSTVEVESVLYGHP 173
Query: 616 IIQQIVVIGQDKRRLG---AIIVPNSEEVLKVARELSIIDSISSNVVSEEKVLNLIYKE 671
+ Q V+ + G V E + + E +++ N+ +++KE
Sbjct: 174 AVNQAAVVARADEFWGETPCAFVSLKEGLKETVTEREVVEFCRKNMPHYMVPKTVVFKE 350
>TC10376 similar to UP|Q8VZF1 (Q8VZF1) AT3g16910/K14A17_3 (Adenosine
monophosphate binding protein 7 AMPBP7), partial (50%)
Length = 888
Score = 35.4 bits (80), Expect = 0.032
Identities = 28/104 (26%), Positives = 48/104 (45%), Gaps = 11/104 (10%)
Frame = +3
Query: 272 YTSGTTGNPKGVMLTHRN--LLHQIKNL-WDTVPAEVGDRFLSMLPPWHAYERACEYFIF 328
YTSGTT +PKGV+L HR L+ L W V +L LP +H C + +
Sbjct: 384 YTSGTTASPKGVVLHHRGAYLMSLCGALVWGMTEGAV---YLWTLPMFH-----CNGWCY 539
Query: 329 TCGIEQVY--------TTVRNLKDDLGRYQPHYMISVPLVFETL 364
T + +Y T + + + + +Y+ + + P+V ++
Sbjct: 540 TWTLPAIYGTNICLRQVTAKAVYEAIAKYKVTHFCAAPVVLNSI 671
>BP053582
Length = 516
Score = 32.0 bits (71), Expect(2) = 0.039
Identities = 21/60 (35%), Positives = 28/60 (46%)
Frame = -1
Query: 536 GPPVMNGYYKNPLATNQALDKDGWLNTGDLGWIAPHHSTGRSRNSSGVIVVDGRAKDTIV 595
G ++N Y KN AT A K GW TGDL ++ G I + R+KD I+
Sbjct: 486 G*TLLNRYLKNLKATQDAF-KGGWFRTGDL----------EVKHPDGYIELKDRSKDLII 340
Score = 21.9 bits (45), Expect(2) = 0.039
Identities = 8/27 (29%), Positives = 14/27 (51%)
Frame = -2
Query: 599 GENVEPAELEEAAMRSSIIQQIVVIGQ 625
GEN+ ELE+ + + V+G+
Sbjct: 326 GENISSIELEDVIFSHPAVLEAAVVGR 246
>BP063946
Length = 392
Score = 33.9 bits (76), Expect = 0.094
Identities = 13/34 (38%), Positives = 23/34 (67%)
Frame = -1
Query: 691 EPFTIDNGLMTPTMKIRRDRVVAKYKEQIDDLYK 724
EP ++ L+TPT +++R ++ YK+ ID LY+
Sbjct: 350 EPLDLERDLITPTFQLKRPPLLQCYKDHIDPLYQ 249
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.319 0.136 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,071,467
Number of Sequences: 28460
Number of extensions: 163358
Number of successful extensions: 937
Number of sequences better than 10.0: 58
Number of HSP's better than 10.0 without gapping: 837
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 896
length of query: 724
length of database: 4,897,600
effective HSP length: 97
effective length of query: 627
effective length of database: 2,136,980
effective search space: 1339886460
effective search space used: 1339886460
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 59 (27.3 bits)
Medicago: description of AC146585.9


