

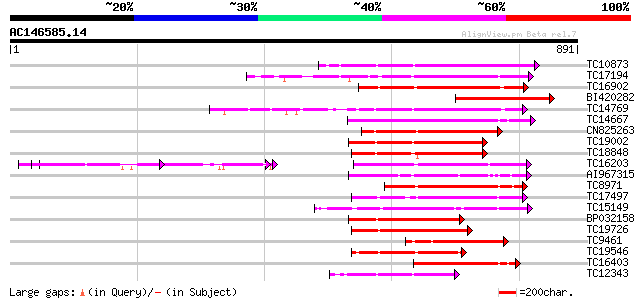
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146585.14 - phase: 0
(891 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC10873 similar to UP|Q9LDZ5 (Q9LDZ5) F2D10.13 (F5M15.3), partia... 235 3e-62
TC17194 UP|CAE02590 (CAE02590) Nod-factor receptor 1b, complete 216 1e-56
TC16902 homologue to UP|Q8VZH5 (Q8VZH5) Receptor protein kinase-... 211 4e-55
BI420282 207 8e-54
TC14769 UP|Q8LKX1 (Q8LKX1) Receptor-like kinase SYMRK, complete 206 2e-53
TC14667 homologue to UP|Q84P43 (Q84P43) Protein kinase Pti1, com... 201 4e-52
CN825263 199 1e-51
TC19002 weakly similar to UP|Q9XJ10 (Q9XJ10) ESTs C22458(C62866)... 195 2e-50
TC18848 weakly similar to UP|Q9SSQ6 (Q9SSQ6) F6D8.24 protein, pa... 193 9e-50
TC16203 UP|Q8GRU6 (Q8GRU6) LRR receptor-like kinase (Hypernodula... 181 6e-46
AI967315 177 5e-45
TC8971 similar to GB|AAM16251.1|20334780|AY093990 At2g11520/F14P... 157 9e-39
TC17497 UP|CAE45595 (CAE45595) S-receptor kinase-like protein 2,... 155 2e-38
TC15149 similar to UP|Q9LVI6 (Q9LVI6) Probable receptor-like pro... 149 2e-36
BP032158 149 2e-36
TC19726 similar to UP|Q9XIS3 (Q9XIS3) Lectin-like protein kinase... 144 8e-35
TC9461 similar to UP|O49840 (O49840) Protein kinase (AT2G02800/T... 142 2e-34
TC19546 similar to UP|Q40543 (Q40543) Protein-serine/threonine k... 142 3e-34
TC16403 homologue to UP|Q9LKY4 (Q9LKY4) Pti1 kinase-like protein... 139 2e-33
TC12343 similar to UP|CDK9_SCHPO (Q96WV9) Serine/threonine prote... 138 4e-33
>TC10873 similar to UP|Q9LDZ5 (Q9LDZ5) F2D10.13 (F5M15.3), partial (77%)
Length = 1478
Score = 235 bits (599), Expect = 3e-62
Identities = 134/349 (38%), Positives = 191/349 (54%), Gaps = 1/349 (0%)
Frame = +1
Query: 485 NGEGNVKLDLASVSNGYGGASSELQSQSSGDHSDLHGFDGGNGGNATISIHVLRQVTNDF 544
+G G V A+ G E + + G S +G G A+ L T +F
Sbjct: 286 SGNGGVNAAAAATD---GERKRETKGKGKGVSSS-NGSSNGKTAAASFGFRELADATRNF 453
Query: 545 SDDNILGRGGFGIVYKGELPDGTKIAVKRMISVAKGSKGLNEFQAEIGVLTKVRHRHLVA 604
+ N++G GGFG VYKG L G +AVK++ G +G EF E+ +L+ + H +LV
Sbjct: 454 KEANLIGEGGFGKVYKGRLTTGEAVAVKQLSH--DGRQGFQEFVMEVLMLSLLHHTNLVR 627
Query: 605 LLGYCINGNERLLVYEHMPQGTLTQHLFECREHGYTPLTWKQRLIIALDVGRGVEYLHSL 664
L+GYC +G++RLLVYE+MP G+L HLFE H PL W R+ +A+ RG+EYLH
Sbjct: 628 LIGYCTDGDQRLLVYEYMPMGSLEDHLFEL-SHDKEPLNWSTRMKVAVGAARGLEYLHCT 804
Query: 665 AQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDG-NYSVETKLAGTFGYLAPEYAAT 723
A I+RDLK +NILL ++ K++DFGL K P G N V T++ GT+GY APEYA +
Sbjct: 805 ADPPVIYRDLKSANILLDNEFNPKLSDFGLAKLGPVGDNTHVSTRVMGTYGYCAPEYAMS 984
Query: 724 GRVTTKVDVYAFGVVLMELITGRKALDDSVPDESSHLVTWFRRVLTNKENIPKAIDQTLD 783
G++T K D+Y+FGVVL+EL+TGR+A+D S +LV+W R +++ +D L
Sbjct: 985 GKLTLKSDIYSFGVVLLELLTGRRAIDTSRRPGEQNLVSWARPYFSDRRRFGHMVDPLLQ 1164
Query: 784 PDEETMLSIYKVAELAGHCTTRSPYQRPDIGHAVNVLCPLVQQWEPTTH 832
+++ + C P RP I V L L Q P H
Sbjct: 1165-GRFPSRCLHQAIAITAMCLQEQPKFRPLITDIVVALEYLASQGSPEAH 1308
>TC17194 UP|CAE02590 (CAE02590) Nod-factor receptor 1b, complete
Length = 2193
Score = 216 bits (551), Expect = 1e-56
Identities = 163/472 (34%), Positives = 234/472 (49%), Gaps = 20/472 (4%)
Frame = +1
Query: 372 GSIPQNLATLSQLETLDVSNNDLSGEVPKFSPKVKFITDGNVWLGKNHGGGAPGSAPGG- 430
G Q++A S L+ +G + F+P V F D + PG G
Sbjct: 634 GDTLQDIANQSSLD---------AGLIQSFNPSVNFSKDSGIAF-------IPGRYKNGV 765
Query: 431 ------SPAGSGKGASMKKVWIIIIIVLIVVGFVVGGAWFSWKCYSRKGLRRFARVGNPE 484
AG GA++ + I VL+++ F + + R E
Sbjct: 766 YVPLYHRTAGLASGAAVG-ISIAGTFVLLLLAFCM-----------------YVRYQKKE 891
Query: 485 NGEGNVKLDLA-SVSNGYGGASSELQSQSSGDHSDLHGFDGGNGGNATI-------SIHV 536
+ + D++ ++S G ASS + ++SG G G + + S
Sbjct: 892 EEKAKLPTDISMALSTQDGNASSSAEYETSGSSGP--GTASATGLTSIMVAKSMEFSYQE 1065
Query: 537 LRQVTNDFSDDNILGRGGFGIVYKGELPDGTKIAVKRMISVAKGSKGLNEFQAEIGVLTK 596
L + TN+FS DN +G+GGFG VY EL G K A+K+M A EF E+ VLT
Sbjct: 1066 LAKATNNFSLDNKIGQGGFGAVYYAELR-GKKTAIKKMDVQAS-----TEFLCELKVLTH 1227
Query: 597 VRHRHLVALLGYCINGNERLLVYEHMPQGTLTQHLFECREHGYTPLTWKQRLIIALDVGR 656
V H +LV L+GYC+ G+ LVYEH+ G L Q+L G PL W R+ IALD R
Sbjct: 1228 VHHLNLVRLIGYCVEGS-LFLVYEHIDNGNLGQYL---HGSGKEPLPWSSRVQIALDAAR 1395
Query: 657 GVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDGNYSVETKLAGTFGYL 716
G+EY+H +IHRD+K +NIL+ ++R KVADFGL K GN +++T+L GTFGY+
Sbjct: 1396 GLEYIHEHTVPVYIHRDVKSANILIDKNLRGKVADFGLTKLIEVGNSTLQTRLVGTFGYM 1575
Query: 717 APEYAATGRVTTKVDVYAFGVVLMELITGRKAL--DDSVPDESSHLVTWFRRVLTNKENI 774
PEYA G ++ K+DVYAFGVVL ELI+ + A+ + ES LV F L NK +
Sbjct: 1576 PPEYAQYGDISPKIDVYAFGVVLFELISAKNAVLKTGELVAESKGLVALFEEAL-NKSDP 1752
Query: 775 PKAIDQTLDP---DEETMLSIYKVAELAGHCTTRSPYQRPDIGHAVNVLCPL 823
A+ + +DP + + S+ K+A+L CT +P RP + V L L
Sbjct: 1753 CDALRKLVDPRLGENYPIDSVLKIAQLGRACTRDNPLLRPSMRSLVVALMTL 1908
>TC16902 homologue to UP|Q8VZH5 (Q8VZH5) Receptor protein kinase-like
protein, partial (46%)
Length = 941
Score = 211 bits (537), Expect = 4e-55
Identities = 119/270 (44%), Positives = 166/270 (61%), Gaps = 4/270 (1%)
Frame = +3
Query: 549 ILGRGGFGIVYKGELPDGTKIAVKRMISVAKGSKGLNEFQAEIGVLTKVRHRHLVALLGY 608
+LG GGFG VY GE+ GTK+A+KR +++ +G++EFQ EI +L+K+RHRHLV+L+GY
Sbjct: 12 LLGVGGFGKVYYGEVDGGTKVAIKRGNPLSE--QGVHEFQTEIEMLSKLRHRHLVSLIGY 185
Query: 609 CINGNERLLVYEHMPQGTLTQHLFECREHGYTPLTWKQRLIIALDVGRGVEYLHSLAQQS 668
C E +LVY+HM GTL +HL++ ++ PL WKQRL I + RG+ YLH+ A+ +
Sbjct: 186 CEENTEMILVYDHMAYGTLREHLYKTQK---PPLPWKQRLEICIGAARGLHYLHTGAKYT 356
Query: 669 FIHRDLKPSNILLGDDMRAKVADFGLVKNAPD-GNYSVETKLAGTFGYLAPEYAATGRVT 727
IHRD+K +NILL + AKV+DFGL K P N V T + G+FGYL PEY ++T
Sbjct: 357 IIHRDVKTTNILLDEKWVAKVSDFGLSKTGPTLDNTHVSTVVKGSFGYLDPEYFRRQQLT 536
Query: 728 TKVDVYAFGVVLMELITGRKALDDSVPDESSHLVTWFRRVLTNKENIPKAIDQTLDPDEE 787
K DVY+FGVVL E++ R AL+ S+ E L W +DQ LDP +
Sbjct: 537 DKSDVYSFGVVLFEILCARPALNPSLAKEQVSLAEWASHCYNK-----GILDQILDPYLK 701
Query: 788 TMLS---IYKVAELAGHCTTRSPYQRPDIG 814
++ K AE A C + +RP +G
Sbjct: 702 GKIAPECFKKFAETAMKCVSDQGIERPSMG 791
>BI420282
Length = 504
Score = 207 bits (526), Expect = 8e-54
Identities = 97/155 (62%), Positives = 123/155 (78%)
Frame = +1
Query: 701 GNYSVETKLAGTFGYLAPEYAATGRVTTKVDVYAFGVVLMELITGRKALDDSVPDESSHL 760
G S ET+LAGTFGYLAPEYA TGRVTTKVDV++FGV+LMEL+TGRKALD++ P++S HL
Sbjct: 37 GKASFETRLAGTFGYLAPEYAVTGRVTTKVDVFSFGVILMELLTGRKALDETQPEDSMHL 216
Query: 761 VTWFRRVLTNKENIPKAIDQTLDPDEETMLSIYKVAELAGHCTTRSPYQRPDIGHAVNVL 820
VTWFRR+ +K++ KAID +++ +EET+ S++ VAELAGHC R PYQRPD+GHAVNVL
Sbjct: 217 VTWFRRMYIDKDSFRKAIDPSIEINEETLASVHTVAELAGHCCAREPYQRPDMGHAVNVL 396
Query: 821 CPLVQQWEPTTHTDESTCADDNQMSLTQALQRWQA 855
LV+ W+P+ E D MSL QAL++WQA
Sbjct: 397 SSLVELWKPSDQNSEDMYGIDLDMSLPQALKKWQA 501
>TC14769 UP|Q8LKX1 (Q8LKX1) Receptor-like kinase SYMRK, complete
Length = 3495
Score = 206 bits (523), Expect = 2e-53
Identities = 162/520 (31%), Positives = 256/520 (49%), Gaps = 21/520 (4%)
Frame = +3
Query: 315 KSWTGNDPC--KDWLCVICGGGK----ITKLNFAKQGLQGTISPAFANLTDLTALYLNGN 368
+SW+G DPC W + C G ITKL+ + L+G I + A +T+L L ++ N
Sbjct: 1521 ESWSG-DPCILLPWKGIACDGSNGSSVITKLDLSSSNLKGLIPSSIAEMTNLETLNISHN 1697
Query: 369 NLTGSIPQNLATLSQLETLDVSNNDLSGEVPKFSPKVKFITDGNVWLGKNHGGGAPGSAP 428
+ GS+P + S L ++D+S NDL G++P+ K+ + +++ G N A
Sbjct: 1698 SFDGSVP-SFPLSSLLISVDLSYNDLMGKLPESIVKLPHLK--SLYFGCNEHMSPEDPAN 1868
Query: 429 GGSPA-----GSGKGASMKKVWIIII------IVLIVVGFVVGGAWFSWKCYSRKGLRRF 477
S G KG + +I+I +LI + F G F + Y +K +
Sbjct: 1869 MNSSLINTDYGRCKGKESRFGQVIVIGAITCGSLLITLAF---GVLFVCR-YRQKLIPWE 2036
Query: 478 ARVGNPENGEGNVKLDLASVSNGYGGASSELQSQSSGDHSDLHGFDGGNGGNATISIHVL 537
G E N+ L S + + ++S S ++ +
Sbjct: 2037 GFAGKKYPMETNIIFSLPSKDDFF------IKSVSI----------------QAFTLEYI 2150
Query: 538 RQVTNDFSDDNILGRGGFGIVYKGELPDGTKIAVKRMISVAKGSKGLNEFQAEIGVLTKV 597
T + ++G GGFG VY+G L DG ++AVK + + ++G EF E+ +L+ +
Sbjct: 2151 EVATERYK--TLIGEGGFGSVYRGTLNDGQEVAVK--VRSSTSTQGTREFDNELNLLSAI 2318
Query: 598 RHRHLVALLGYCINGNERLLVYEHMPQGTLTQHLFECREHGYTPLTWKQRLIIALDVGRG 657
+H +LV LLGYC ++++LVY M G+L L+ L W RL IAL RG
Sbjct: 2319 QHENLVPLLGYCNESDQQILVYPFMSNGSLQDRLYG-EPAKRKILDWPTRLSIALGAARG 2495
Query: 658 VEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAP-DGNYSVETKLAGTFGYL 716
+ YLH+ +S IHRD+K SNILL M AKVADFG K AP +G+ V ++ GT GYL
Sbjct: 2496 LAYLHTFPGRSVIHRDIKSSNILLDHSMCAKVADFGFSKYAPQEGDSYVSLEVRGTAGYL 2675
Query: 717 APEYAATGRVTTKVDVYAFGVVLMELITGRKALDDSVPDESSHLVTWFRRVLTNKENIPK 776
PEY T +++ K DV++FGVVL+E+++GR+ L+ P LV W + +
Sbjct: 2676 DPEYYKTQQLSEKSDVFSFGVVLLEIVSGREPLNIKRPRTEWSLVEWATPYIRGSK---- 2843
Query: 777 AIDQTLDPDEE---TMLSIYKVAELAGHCTTRSPYQRPDI 813
+D+ +DP + ++++V E+A C RP +
Sbjct: 2844 -VDEIVDPGIKGGYHAEAMWRVVEVALQCLEPFSTYRPSM 2960
Score = 37.0 bits (84), Expect = 0.014
Identities = 24/86 (27%), Positives = 45/86 (51%), Gaps = 1/86 (1%)
Frame = +3
Query: 80 SSLTDVNLGSNNFSSVTPGAFSGLNSLQTLSLGEN-INLSPWTFPTELTQSSNLNSIDIN 138
S +T ++L S+N + P + + + +L+TL++ N + S +FP SS L S+D++
Sbjct: 1593 SVITKLDLSSSNLKGLIPSSIAEMTNLETLNISHNSFDGSVPSFPL----SSLLISVDLS 1760
Query: 139 QAKINGTLPDIFGSFSSLNTLHLAYN 164
+ G LP+ L +L+ N
Sbjct: 1761 YNDLMGKLPESIVKLPHLKSLYFGCN 1838
>TC14667 homologue to UP|Q84P43 (Q84P43) Protein kinase Pti1, complete
Length = 1591
Score = 201 bits (511), Expect = 4e-52
Identities = 115/299 (38%), Positives = 178/299 (59%), Gaps = 5/299 (1%)
Frame = +2
Query: 532 ISIHVLRQVTNDFSDDNILGRGGFGIVYKGELPDGTKIAVKRMISVAKGSKGLNEFQAEI 591
+S+ L++ T++F ++G G +G VY L DG +AVK++ V+ + NEF ++
Sbjct: 323 LSLDELKEKTDNFGSKALIGEGSYGRVYYATLNDGNAVAVKKL-DVSSEPETNNEFLTQV 499
Query: 592 GVLTKVRHRHLVALLGYCINGNERLLVYEHMPQGTLTQHLFECRE-HGYTP---LTWKQR 647
+++++++ + V L GYC+ GN R+L YE G+L L + G P L W QR
Sbjct: 500 SMVSRLKNDNFVELHGYCVEGNLRVLAYEFATMGSLHDILHGRKGVQGAQPGPTLDWIQR 679
Query: 648 LIIALDVGRGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDGNYSVE- 706
+ IA+D RG+EYLH Q + IHRD++ SN+L+ +D +AK+ADF L APD +
Sbjct: 680 VRIAVDAARGLEYLHEKVQPAIIHRDIRSSNVLIFEDYKAKIADFNLSNQAPDMAARLHS 859
Query: 707 TKLAGTFGYLAPEYAATGRVTTKVDVYAFGVVLMELITGRKALDDSVPDESSHLVTWFRR 766
T++ GTFGY APEYA TG++T K DVY+FGVVL+EL+TGRK +D ++P LVTW
Sbjct: 860 TRVLGTFGYHAPEYAMTGQLTQKSDVYSFGVVLLELLTGRKPVDHTMPRGQQSLVTWATP 1039
Query: 767 VLTNKENIPKAIDQTLDPDEETMLSIYKVAELAGHCTTRSPYQRPDIGHAVNVLCPLVQ 825
L +++ + + +D L E + K+A +A C RP++ V L PL++
Sbjct: 1040RL-SEDKVKQCVDPKL-KGEYPPKGVAKLAAVAALCVQYEAEFRPNMSIVVKALQPLLK 1210
>CN825263
Length = 663
Score = 199 bits (507), Expect = 1e-51
Identities = 110/223 (49%), Positives = 142/223 (63%), Gaps = 2/223 (0%)
Frame = +1
Query: 554 GFGIVYKGELPDGTKIAVKRMISVAKGSKGLNEFQAEIGVLTKVRHRHLVALLGYCINGN 613
GFG+VYKG L DG +AVK I +G EF AE+ +L+++ HR+LV L+G CI
Sbjct: 1 GFGLVYKGILNDGRDVAVK--ILKRDDQRGGREFLAEVEMLSRLHHRNLVKLIGICIEKQ 174
Query: 614 ERLLVYEHMPQGTLTQHLFEC-REHGYTPLTWKQRLIIALDVGRGVEYLHSLAQQSFIHR 672
R L+YE +P G++ HL +E G PL W R+ IAL RG+ YLH + IHR
Sbjct: 175 TRCLIYELVPNGSVESHLHGADKETG--PLDWNARMKIALGAARGLAYLHEDSNPCVIHR 348
Query: 673 DLKPSNILLGDDMRAKVADFGLVKNAPD-GNYSVETKLAGTFGYLAPEYAATGRVTTKVD 731
D K SNILL D KV+DFGL + A D GN + T + GTFGYLAPEYA TG + K D
Sbjct: 349 DFKSSNILLECDFTPKVSDFGLARTALDEGNKHISTHVMGTFGYLAPEYAMTGHLLVKSD 528
Query: 732 VYAFGVVLMELITGRKALDDSVPDESSHLVTWFRRVLTNKENI 774
VY++GVVL+EL+TG K +D S P +LVTW R +LT+KE +
Sbjct: 529 VYSYGVVLLELLTGTKPVDLSQPPGQENLVTWARPILTSKEGL 657
>TC19002 weakly similar to UP|Q9XJ10 (Q9XJ10) ESTs C22458(C62866), partial
(48%)
Length = 780
Score = 195 bits (496), Expect = 2e-50
Identities = 101/218 (46%), Positives = 140/218 (63%)
Frame = +1
Query: 533 SIHVLRQVTNDFSDDNILGRGGFGIVYKGELPDGTKIAVKRMISVAKGSKGLNEFQAEIG 592
S+ L TN+F+ DN LG GGFG VY G+L DG++IAVKR+ +K EF E+
Sbjct: 31 SLKELHSATNNFNYDNKLGEGGFGSVYWGQLWDGSQIAVKRL--KVWSNKADMEFAVEVE 204
Query: 593 VLTKVRHRHLVALLGYCINGNERLLVYEHMPQGTLTQHLFECREHGYTPLTWKQRLIIAL 652
+L +VRH++L++L GYC G ERL+VY++MP +L HL + L W +R+ IA+
Sbjct: 205 ILARVRHKNLLSLRGYCAEGQERLIVYDYMPNLSLLSHLHG-QHSSECLLDWNRRMNIAI 381
Query: 653 DVGRGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDGNYSVETKLAGT 712
G+ YLH A IHRD+K SN+LL D +A+VADFG K PDG V T++ GT
Sbjct: 382 GSAEGIVYLHHQATPHIIHRDIKASNVLLDSDFQARVADFGFAKLIPDGATHVTTRVKGT 561
Query: 713 FGYLAPEYAATGRVTTKVDVYAFGVVLMELITGRKALD 750
GYLAPEYA G+ DV++FG++L+EL +G+K L+
Sbjct: 562 LGYLAPEYAMLGKANECCDVFSFGILLLELASGKKPLE 675
>TC18848 weakly similar to UP|Q9SSQ6 (Q9SSQ6) F6D8.24 protein, partial (64%)
Length = 969
Score = 193 bits (491), Expect = 9e-50
Identities = 101/218 (46%), Positives = 136/218 (62%), Gaps = 4/218 (1%)
Frame = +2
Query: 537 LRQVTNDFSDDNILGRGGFGIVYKGELPDGTKIAVKRMISVAKGSKGLNEFQAEIGVLTK 596
L T FSDDN LG GGFG VY G DG +IAVK++ A SK EF E+ VL +
Sbjct: 281 LHAATGGFSDDNKLGEGGFGSVYWGRTSDGLQIAVKKL--KAMNSKAEMEFAVEVEVLGR 454
Query: 597 VRHRHLVALLGYCINGNERLLVYEHMPQGTLTQHLFECREHGY----TPLTWKQRLIIAL 652
VRH++L+ L GYC+ ++RL+VY++MP +L HL HG L W++R+ IA+
Sbjct: 455 VRHKNLLGLRGYCVGDDQRLIVYDYMPNLSLLSHL-----HGQFAVEVQLNWQKRMKIAI 619
Query: 653 DVGRGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDGNYSVETKLAGT 712
G+ YLH IHRD+K SN+LL D VADFG K P+G + T++ GT
Sbjct: 620 GSAEGILYLHHEVTPHIIHRDIKASNVLLNSDFEPLVADFGFAKLIPEGVSHMTTRVKGT 799
Query: 713 FGYLAPEYAATGRVTTKVDVYAFGVVLMELITGRKALD 750
GYLAPEYA G+V+ DVY+FG++L+EL+TGRK ++
Sbjct: 800 LGYLAPEYAMWGKVSESCDVYSFGILLLELVTGRKPIE 913
>TC16203 UP|Q8GRU6 (Q8GRU6) LRR receptor-like kinase (Hypernodulation aberrant
root formation protein), complete
Length = 3308
Score = 181 bits (458), Expect = 6e-46
Identities = 108/286 (37%), Positives = 165/286 (56%), Gaps = 5/286 (1%)
Frame = +2
Query: 540 VTNDFSDDNILGRGGFGIVYKGELPDGTKIAVKRMISVAKGSKGLNEFQAEIGVLTKVRH 599
V ++NI+G+GG GIVY+G +P+GT +A+KR++ G F+AEI L K+RH
Sbjct: 2180 VVECLKEENIIGKGGAGIVYRGSMPNGTDVAIKRLVGQGSGRNDYG-FRAEIETLGKIRH 2356
Query: 600 RHLVALLGYCINGNERLLVYEHMPQGTLTQHLFECREHGYTPLTWKQRLIIALDVGRGVE 659
R+++ LLGY N + LL+YE+MP G+L + L + G+ L W+ R IA++ RG+
Sbjct: 2357 RNIMRLLGYVSNKDTNLLLYEYMPNGSLGEWLHGAKG-GH--LRWEMRYKIAVEAARGLC 2527
Query: 660 YLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDGNYSVE-TKLAGTFGYLAP 718
Y+H IHRD+K +NILL D A VADFGL K D S + +AG++GY+AP
Sbjct: 2528 YMHHDCSPLIIHRDVKSNNILLDADFEAHVADFGLAKFLYDPGASQSMSSIAGSYGYIAP 2707
Query: 719 EYAATGRVTTKVDVYAFGVVLMELITGRKALDDSVPDESSHLVTWFRRVLT--NKENIPK 776
EYA T +V K DVY+FGVVL+ELI GRK + + + +V W + ++ ++ +
Sbjct: 2708 EYAYTLKVDEKSDVYSFGVVLLELIIGRKPVGEF--GDGVDIVGWVNKTMSELSQPSDTA 2881
Query: 777 AIDQTLDP--DEETMLSIYKVAELAGHCTTRSPYQRPDIGHAVNVL 820
+ +DP + S+ + +A C RP + V++L
Sbjct: 2882 LVLAVVDPRLSGYPLTSVIHMFNIAMMCVKEMGPARPTMREVVHML 3019
Score = 136 bits (342), Expect = 2e-32
Identities = 138/468 (29%), Positives = 209/468 (44%), Gaps = 61/468 (13%)
Frame = +2
Query: 15 WSSNTSF---CQWSGVKCSSDNRVTSINLSDQKLAGTLPDNLNSLTQLTTLYLQNNALSG 71
W +TS C +SGV C + RV ++N++ L G LP + L +L L + N L+
Sbjct: 266 WKFSTSLSAHCSFSGVTCDQNLRVVALNVTLVPLFGHLPPEIGLLEKLENLTISMNNLTD 445
Query: 72 PLPS-LANLSSLTDVNLGSNNFSSVTPGAFS-GLNSLQTLSLGENINLSPWTFPTELTQS 129
LPS LA+L+SL +N+ N FS PG + G+ L+ L +N P P E+ +
Sbjct: 446 QLPSDLASLTSLKVLNISHNLFSGQFPGNITVGMTELEALDAYDNSFSGP--LPEEIVKL 619
Query: 130 SNLNSIDINQAKINGTLPDIFGSFSSLNTLHLAYNNLSGGLPNSLA-------------- 175
L + + +GT+P+ + F SL L L N+L+G +P SLA
Sbjct: 620 EKLKYLHLAGNYFSGTIPESYSEFQSLEFLGLNANSLTGRVPESLAKLKTLKELHLGYSN 799
Query: 176 ---GSGIQSFWINNNLP-------GLTGSI-TVISNMTLLTQVWLHVNKFTGPI-PDLSQ 223
G +F NL LTG I + N+T L +++ +N TG I P+LS
Sbjct: 800 AYEGGIPPAFGSMENLRLLEMANCNLTGEIPPSLGNLTKLHSLFVQMNNLTGTIPPELSS 979
Query: 224 CNSIKDLQLRDNQLTGVVPDSLVSMSGLQNVTLRN---NQLQGPVPVFGKDV-KYNSDDI 279
S+ L L N LTG +P+ S S L+N+TL N N+ +G +P F D+ + +
Sbjct: 980 MMSLMSLDLSINDLTGEIPE---SFSKLKNLTLMNFFQNKFRGSLPSFIGDLPNLETLQV 1150
Query: 280 SHNNFCNNNASVPCDARVMDLLHIVGGFGYPIQF---AKSWTG---NDPCKDWLC----- 328
NNF L H +GG G + F TG D CK
Sbjct: 1151WENNFS------------FVLPHNLGGNGRFLYFDVTKNHLTGLIPPDLCKSGRLKTFII 1294
Query: 329 --------VICGGGK---ITKLNFAKQGLQGTISPAFANLTDLTALYLNGNNLTGSIPQN 377
+ G G+ +TK+ A L G + P L +T L+ N L G +P
Sbjct: 1295TDNFFRGPIPKGIGECRSLTKIRVANNFLDGPVPPGVFQLPSVTITELSNNRLNGELPSV 1474
Query: 378 LATLSQLETLDVSNNDLSGEVPKFSPKVKFI----TDGNVWLGKNHGG 421
++ S L TL +SNN +G++P ++ + D N ++G+ GG
Sbjct: 1475ISGES-LGTLTLSNNLFTGKIPAAMKNLRALQSLSLDANEFIGEIPGG 1615
Score = 127 bits (320), Expect = 6e-30
Identities = 105/372 (28%), Positives = 168/372 (44%), Gaps = 9/372 (2%)
Frame = +2
Query: 47 GTLPDNLNSLTQLTTLYLQNNALSGPLP-SLANLSSLTDVNLGSNNFSSVTPGAFSGLNS 105
G +P S+ L L + N L+G +P SL NL+ L + + NN + P S + S
Sbjct: 809 GGIPPAFGSMENLRLLEMANCNLTGEIPPSLGNLTKLHSLFVQMNNLTGTIPPELSSMMS 988
Query: 106 LQTLSLGENINLSPWTFPTELTQSSNLNSIDINQAKINGTLPDIFGSFSSLNTLHLAYNN 165
L +L L +IN P ++ NL ++ Q K G+LP G +L TL + NN
Sbjct: 989 LMSLDL--SINDLTGEIPESFSKLKNLTLMNFFQNKFRGSLPSFIGDLPNLETLQVWENN 1162
Query: 166 LSGGLPNSLAGSGIQSFW--INNNLPGLTGSITVISNMTLLTQVWLHVNKFTGPIPD-LS 222
S LP++L G+G ++ N+L GL S L + N F GPIP +
Sbjct: 1163 FSFVLPHNLGGNGRFLYFDVTKNHLTGLIPPDLCKSGR--LKTFIITDNFFRGPIPKGIG 1336
Query: 223 QCNSIKDLQLRDNQLTGVVPDSLVSMSGLQNVTLRNNQLQGPVPVFGKDVKYNSDDISHN 282
+C S+ +++ +N L G VP + + + L NN+L G +P + +S+N
Sbjct: 1337 ECRSLTKIRVANNFLDGPVPPGVFQLPSVTITELSNNRLNGELPSVISGESLGTLTLSNN 1516
Query: 283 NFCNNNASVPCDARVMDLLHIVGGFGYPIQFAKSWTGNDPCKDWLCVICGGG-----KIT 337
F + + R + L + A + G P GG +T
Sbjct: 1517 LFTGKIPAAMKNLRALQSLSLD---------ANEFIGEIP----------GGVFEIPMLT 1639
Query: 338 KLNFAKQGLQGTISPAFANLTDLTALYLNGNNLTGSIPQNLATLSQLETLDVSNNDLSGE 397
K+N + L G I + LTA+ L+ NNL G +P+ + L L L++S N++SG
Sbjct: 1640 KVNISGNNLTGPIPTTITHRASLTAVDLSRNNLAGEVPKGMKNLMDLSILNLSRNEISGP 1819
Query: 398 VPKFSPKVKFIT 409
VP +++F+T
Sbjct: 1820 VP---DEIRFMT 1846
Score = 79.0 bits (193), Expect = 3e-15
Identities = 58/209 (27%), Positives = 95/209 (44%), Gaps = 1/209 (0%)
Frame = +2
Query: 35 VTSINLSDQKLAGTLPDNLNSLTQLTTLYLQNNALSGPLPSLANLSSLTDVNLGSNNFSS 94
+T I +++ L G +P + L +T L NN L+G LPS+ + SL + L +N F+
Sbjct: 1349 LTKIRVANNFLDGPVPPGVFQLPSVTITELSNNRLNGELPSVISGESLGTLTLSNNLFTG 1528
Query: 95 VTPGAFSGLNSLQTLSLGENINLSPWTFPTELTQSSNLNSIDINQAKINGTLPDIFGSFS 154
P A L +LQ+LSL N + P + + L ++I+ + G +P +
Sbjct: 1529 KIPAAMKNLRALQSLSLDANEFIG--EIPGGVFEIPMLTKVNISGNNLTGPIPTTITHRA 1702
Query: 155 SLNTLHLAYNNLSGGLPNSLAGSGIQSFWINNNLPGLTGSITVISNMTLLTQVWLHVNKF 214
SL + L+ NNL+G +P + N+ L+ + L N+
Sbjct: 1703 SLTAVDLSRNNLAGEVPKGM------------------------KNLMDLSILNLSRNEI 1810
Query: 215 TGPIPD-LSQCNSIKDLQLRDNQLTGVVP 242
+GP+PD + S+ L L N TG VP
Sbjct: 1811 SGPVPDEIRFMTSLTTLDLSSNNFTGTVP 1897
Score = 36.6 bits (83), Expect = 0.018
Identities = 15/38 (39%), Positives = 26/38 (67%)
Frame = +2
Query: 38 INLSDQKLAGTLPDNLNSLTQLTTLYLQNNALSGPLPS 75
+NLS +++G +PD + +T LTTL L +N +G +P+
Sbjct: 1787 LNLSRNEISGPVPDEIRFMTSLTTLDLSSNNFTGTVPT 1900
>AI967315
Length = 1308
Score = 177 bits (450), Expect = 5e-45
Identities = 113/290 (38%), Positives = 161/290 (54%), Gaps = 2/290 (0%)
Frame = +1
Query: 533 SIHVLRQVTNDFSDDNILGRGGFGIVYKGELPDGTKIAVKRMISVAKGSKGLNEFQAEIG 592
S L TN FS +N++G+GG+ VYKG L G +IAVKR+ + + EF EIG
Sbjct: 115 SYEELFHATNGFSSENMVGKGGYAEVYKGRLESGDEIAVKRLTRTCRDERKEKEFLTEIG 294
Query: 593 VLTKVRHRHLVALLGYCINGNERLLVYEHMPQGTLTQHLFECREHGYTPLTWKQRLIIAL 652
+ V H +++ LLG CI+ N LV+E G++ + + PL WK R I L
Sbjct: 295 TIGHVCHSNVMPLLGCCID-NGLYLVFELSTVGSVASLI---HDEKMAPLDWKTRYKIVL 462
Query: 653 DVGRGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPD--GNYSVETKLA 710
RG+ YLH Q+ IHRD+K SNILL +D +++DFGL K P ++S+ +
Sbjct: 463 GTARGLHYLHKGCQRRIIHRDIKASNILLTEDFEPQISDFGLAKWLPSQWTHHSI-APIE 639
Query: 711 GTFGYLAPEYAATGRVTTKVDVYAFGVVLMELITGRKALDDSVPDESSHLVTWFRRVLTN 770
GTFG+LAPEY G V K DV+AFGV L+E+I+GRK +D S +S H TW + +L +
Sbjct: 640 GTFGHLAPEYYMHGVVDEKTDVFAFGVFLLEVISGRKPVDGS--HQSLH--TWAKPIL-S 804
Query: 771 KENIPKAIDQTLDPDEETMLSIYKVAELAGHCTTRSPYQRPDIGHAVNVL 820
K I K +D L+ + + +VA A C S RP + + V+
Sbjct: 805 KWEIEKLVDPRLEGCYD-VTQFNRVAFAASLCIRASSTWRPTMSEVLEVM 951
>TC8971 similar to GB|AAM16251.1|20334780|AY093990 At2g11520/F14P14.15
{Arabidopsis thaliana;}, partial (37%)
Length = 1079
Score = 157 bits (396), Expect = 9e-39
Identities = 92/227 (40%), Positives = 141/227 (61%), Gaps = 3/227 (1%)
Frame = +3
Query: 590 EIGVLTKVRHRHLVALLGYCINGNERLLVYEHMPQGTLTQHLFECREHGYTPLTWKQRLI 649
E+ +L K+ HR+LV LLG+ GNER+L+ E++P GTL +HL R L + QRL
Sbjct: 6 EVELLAKIDHRNLVKLLGFIDKGNERILITEYVPNGTLREHLDGLRGK---ILDFNQRLE 176
Query: 650 IALDVGRGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAP-DGNYS-VET 707
IA+DV G+ YLH A++ IHRD+K SNILL + MRAKVADFG + P +G+ + + T
Sbjct: 177 IAIDVAHGLTYLHLYAEKQIIHRDVKSSNILLTESMRAKVADFGFARLGPVNGDQTHIST 356
Query: 708 KLAGTFGYLAPEYAATGRVTTKVDVYAFGVVLMELITGRKALD-DSVPDESSHLVTWFRR 766
K+ GT GYL PEY T ++T K DVY+FG++L+E++TGR+ ++ DE L FR+
Sbjct: 357 KVKGTVGYLDPEYMKTHQLTPKSDVYSFGILLLEILTGRRPVELKKAADERVTLRWAFRK 536
Query: 767 VLTNKENIPKAIDQTLDPDEETMLSIYKVAELAGHCTTRSPYQRPDI 813
N+ ++ + +D ++ + + K+ +L+ C RP++
Sbjct: 537 Y--NEGSVVELMDPLMEEAVNADV-LMKMLDLSFQCAAPIRTDRPNM 668
>TC17497 UP|CAE45595 (CAE45595) S-receptor kinase-like protein 2, complete
Length = 2946
Score = 155 bits (393), Expect = 2e-38
Identities = 101/278 (36%), Positives = 154/278 (55%), Gaps = 1/278 (0%)
Frame = +1
Query: 537 LRQVTNDFSDDNILGRGGFGIVYKGELPDGTKIAVKRMISVAKGSKGLNEFQAEIGVLTK 596
+ TN FS N LG GGFG VYKG L +G +IAVKR+ + + +G+ EF+ EI ++ +
Sbjct: 1651 ISSATNHFSLSNKLGEGGFGPVYKGLLANGQEIAVKRLSNTS--GQGMEEFKNEIKLIAR 1824
Query: 597 VRHRHLVALLGYCINGNERLLVYEHMPQGTLTQHLFECREHGYTPLTWKQRLIIALDVGR 656
++HR+LV L G ++ +E + M + L + + W +RL I + R
Sbjct: 1825 LQHRNLVKLFGCSVHQDENSHANKKM------KILLDSTRSKL--VDWNKRLQIIDGIAR 1980
Query: 657 GVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDGNYSVETK-LAGTFGY 715
G+ YLH ++ IHRDLK SNILL D+M K++DFGL + TK + GT+GY
Sbjct: 1981 GLLYLHQDSRLRIIHRDLKTSNILLDDEMNPKISDFGLARIFIGDQVEARTKRVMGTYGY 2160
Query: 716 LAPEYAATGRVTTKVDVYAFGVVLMELITGRKALDDSVPDESSHLVTWFRRVLTNKENIP 775
+ PEYA G + K DV++FGV+++E+I+G+K P +L++ R L +E
Sbjct: 2161 MPPEYAVHGSFSIKSDVFSFGVIVLEIISGKKIGRFYDPHHHLNLLSHAWR-LWIEERPL 2337
Query: 776 KAIDQTLDPDEETMLSIYKVAELAGHCTTRSPYQRPDI 813
+ +D+ LD D I + +A C R P RPD+
Sbjct: 2338 ELVDELLD-DPVIPTEILRYIHVALLCVQRRPENRPDM 2448
>TC15149 similar to UP|Q9LVI6 (Q9LVI6) Probable receptor-like protein kinase
protein (AT3g17840/MEB5_6), partial (45%)
Length = 1489
Score = 149 bits (376), Expect = 2e-36
Identities = 114/345 (33%), Positives = 166/345 (48%), Gaps = 3/345 (0%)
Frame = +3
Query: 480 VGNPENGEGNVKLDLASVSNGYGGASSELQSQSSGDHSDLHGFDGGNGGNATISIHVLRQ 539
+GN NG GN NGY A++ + + GN +LR
Sbjct: 129 IGNG-NGNGN--------GNGYSAAAATINKVEANGVGAKKLVFFGNSARGFDLEDLLR- 278
Query: 540 VTNDFSDDNILGRGGFGIVYKGELPDGTKIAVKRMISVAKGSKGLNEFQAEIGVLTKVRH 599
+ +LG+G FG YK L G +AVKR+ V K EF+ +I + + H
Sbjct: 279 -----ASAEVLGKGTFGTAYKAVLETGLVVAVKRLKDVTISEK---EFKDKIETVGAMDH 434
Query: 600 RHLVALLGYCINGNERLLVYEHMPQGTLTQHLFECREHGYTPLTWKQRLIIALDVGRGVE 659
+ LV L Y + +E+LLVY++MP G+L+ L + G TPL W+ R IAL RG+E
Sbjct: 435 QSLVPLRAYYFSRDEKLLVYDYMPMGSLSALLHGNKGAGRTPLNWEIRSGIALGAARGIE 614
Query: 660 YLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDGNYSVETKLAGTFGYLAPE 719
YLHS + H ++K SNILL AKV+DFGL G S ++A GY APE
Sbjct: 615 YLHSQG-PNVSHGNIKASNILLTKSYEAKVSDFGLAHLV--GPSSTPNRVA---GYRAPE 776
Query: 720 YAATGRVTTKVDVYAFGVVLMELITGRKALDDSVPDESSHLVTWFRRVLTNKENIPKAID 779
+V+ K DVY+FGV+L+EL+TG+ + +E L W + L +E + D
Sbjct: 777 VTDPRKVSQKADVYSFGVLLLELLTGKAPTHALLNEEGVDLPRWVQS-LVREEWTSEVFD 953
Query: 780 QTLDPDEETMLSIYKVAELAGHCTTRSPYQRPDIG---HAVNVLC 821
L + + ++ +LA C P +RP I ++ LC
Sbjct: 954 LELLRYQNVEEEMVQLLQLAVDCAAPYPDRRPSIAEVTRSIEELC 1088
>BP032158
Length = 555
Score = 149 bits (376), Expect = 2e-36
Identities = 76/182 (41%), Positives = 115/182 (62%)
Frame = +2
Query: 533 SIHVLRQVTNDFSDDNILGRGGFGIVYKGELPDGTKIAVKRMISVAKGSKGLNEFQAEIG 592
S+ ++ TN+F N +G GGFG VYKG L +G IAVK++ S K +G EF EIG
Sbjct: 17 SLRQIKAATNNFDPANKIGEGGFGPVYKGVLSEGDVIAVKQLSS--KSKQGNREFINEIG 190
Query: 593 VLTKVRHRHLVALLGYCINGNERLLVYEHMPQGTLTQHLFECREHGYTPLTWKQRLIIAL 652
+++ ++H +LV L G CI GN+ LLVYE+M +L + LF E L W+ R+ I +
Sbjct: 191 MISALQHPNLVKLYGCCIEGNQLLLVYEYMENNSLARALFGNEEQKLN-LNWRTRMKICV 367
Query: 653 DVGRGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDGNYSVETKLAGT 712
+ +G+ YLH ++ +HRD+K +N+LL D++AK++DFGL K + N + T++AGT
Sbjct: 368 GIAKGLAYLHEESRLKIVHRDIKATNVLLDKDLKAKISDFGLAKLDEEENTHISTRIAGT 547
Query: 713 FG 714
G
Sbjct: 548 IG 553
>TC19726 similar to UP|Q9XIS3 (Q9XIS3) Lectin-like protein kinase, partial
(30%)
Length = 728
Score = 144 bits (362), Expect = 8e-35
Identities = 81/196 (41%), Positives = 121/196 (61%), Gaps = 5/196 (2%)
Frame = +3
Query: 537 LRQVTNDFSDDNILGRGGFGIVYKGELP-DGTKIAVKRMISVAKGSKGLNEFQAEIGVLT 595
L++ TN+F + + LG+GG+G+VY+G LP + ++AVK M S K K ++F +E+ ++
Sbjct: 147 LKKATNNFDEKHKLGQGGYGVVYRGMLPKEKLEVAVK-MFSRDK-MKSTDDFLSELIIIN 320
Query: 596 KVRHRHLVALLGYCINGNERLLVYEHMPQGTLTQHLFECREHGY--TPLTWKQRLIIALD 653
++RH+HLV L G+C LLVY++MP G+L H+F C E G TPL+W R I
Sbjct: 321 RLRHKHLVRLQGWCHKNGVLLLVYDYMPNGSLDSHIF-CEEGGTITTPLSWPLRYKIISG 497
Query: 654 VGRGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDG--NYSVETKLAG 711
V + YLH+ Q +HRDLK SNI+L + AK+ DFGL + + +Y+ + G
Sbjct: 498 VASALNYLHNEYDQKVVHRDLKASNIMLDSEFNAKLGDFGLARALENEKISYTELEGVHG 677
Query: 712 TFGYLAPEYAATGRVT 727
T GY+APE TG+ T
Sbjct: 678 TMGYIAPECFHTGKAT 725
>TC9461 similar to UP|O49840 (O49840) Protein kinase (AT2G02800/T20F6.6) ,
partial (37%)
Length = 484
Score = 142 bits (359), Expect = 2e-34
Identities = 74/163 (45%), Positives = 107/163 (65%), Gaps = 1/163 (0%)
Frame = +3
Query: 622 MPQGTLTQHLFECREHGYTPLTWKQRLIIALDVGRGVEYLHSLAQQSFIHRDLKPSNILL 681
MP+G+L HLF G PL+W R+ +A+ RG+ +LH+ A+ I+RD K SNILL
Sbjct: 6 MPKGSLENHLFR---RGPQPLSWSVRMKVAIGAARGLSFLHN-AKSQVIYRDFKASNILL 173
Query: 682 GDDMRAKVADFGLVKNAPDGNYS-VETKLAGTFGYLAPEYAATGRVTTKVDVYAFGVVLM 740
+ AK++DFGL K P G+ + V T++ GT GY APEY ATGR+T K DVY+FGVVL+
Sbjct: 174 DAEFNAKLSDFGLAKAGPTGDRTHVSTQVMGTQGYAAPEYVATGRLTAKSDVYSFGVVLL 353
Query: 741 ELITGRKALDDSVPDESSHLVTWFRRVLTNKENIPKAIDQTLD 783
EL++GR+A+D ++ +LV W R L +K + + +D L+
Sbjct: 354 ELLSGRRAVDKTIAGVDQNLVDWARPYLGDKRRLFRIMDSKLE 482
>TC19546 similar to UP|Q40543 (Q40543) Protein-serine/threonine kinase ,
partial (46%)
Length = 731
Score = 142 bits (357), Expect = 3e-34
Identities = 81/183 (44%), Positives = 116/183 (63%), Gaps = 1/183 (0%)
Frame = +1
Query: 537 LRQVTNDFSDDNILGRGGFGIVYKGELPDGTKIAVKRMISVAKGSKGLNEFQAEIGVLTK 596
+++ T +F+ ILG+G FG VYK +P G +AVK + +K +G +EFQ E+ +L +
Sbjct: 214 IQKATQNFT--TILGQGSFGTVYKATMPTGEVVAVKVLAPNSK--QGEHEFQTEVHLLGR 381
Query: 597 VRHRHLVALLGYCINGNERLLVYEHMPQGTLTQHLFECREHGYTPLTWKQRLIIALDVGR 656
+ HR+LV L+G+C++ +R+LVY+ M G+L L+ + L+W +RL IA+D+
Sbjct: 382 LHHRNLVNLVGFCVDKGQRILVYQFMSNGSLANLLYGEEKE----LSWDERLQIAMDISH 549
Query: 657 GVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAP-DGNYSVETKLAGTFGY 715
G+EYLH A IHRDLK +NILL D MRA VADFGL K DG S L GT+GY
Sbjct: 550 GIEYLHEGAVPPVIHRDLKSANILLDDSMRAMVADFGLSKEEIFDGRNS---GLKGTYGY 720
Query: 716 LAP 718
+ P
Sbjct: 721 MDP 729
>TC16403 homologue to UP|Q9LKY4 (Q9LKY4) Pti1 kinase-like protein, partial
(55%)
Length = 876
Score = 139 bits (351), Expect = 2e-33
Identities = 80/172 (46%), Positives = 109/172 (62%), Gaps = 4/172 (2%)
Frame = +3
Query: 635 REHGYTP---LTWKQRLIIALDVGRGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVAD 691
R G P L+W QR+ IA+ RG+EYLH A+ IHR +K SNILL DD AK+AD
Sbjct: 6 RHEGAQPGLVLSWTQRVKIAVGAARGLEYLHEKAETHIIHRYIKSSNILLFDDDVAKIAD 185
Query: 692 FGLVKNAPDGNYSVE-TKLAGTFGYLAPEYAATGRVTTKVDVYAFGVVLMELITGRKALD 750
F L APD + T++ GTFGY APEYA TG++T+K DVY+FGVVL+EL+TGRK +D
Sbjct: 186 FDLSNQAPDAAARLHSTRVLGTFGYHAPEYAMTGQLTSKSDVYSFGVVLLELLTGRKPVD 365
Query: 751 DSVPDESSHLVTWFRRVLTNKENIPKAIDQTLDPDEETMLSIYKVAELAGHC 802
++P LVTW L +++ + + +D L E + S+ K+A +A C
Sbjct: 366 HTLPRGQQSLVTWATPKL-SEDKVKQCVDARL-KGEYPVKSVAKMAAVAALC 515
>TC12343 similar to UP|CDK9_SCHPO (Q96WV9) Serine/threonine protein kinase
Cdk9 (Cyclin-dependent kinase Cdk9) , partial (7%)
Length = 723
Score = 138 bits (347), Expect = 4e-33
Identities = 82/205 (40%), Positives = 120/205 (58%)
Frame = +3
Query: 503 GASSELQSQSSGDHSDLHGFDGGNGGNATISIHVLRQVTNDFSDDNILGRGGFGIVYKGE 562
G+SSE Q++ D+ + T S L T +F N LG GGFG V+KG+
Sbjct: 141 GSSSEAQNED-----DIQNIAAKE--HKTFSYETLVAATKNFHAVNKLGEGGFGPVFKGK 299
Query: 563 LPDGTKIAVKRMISVAKGSKGLNEFQAEIGVLTKVRHRHLVALLGYCINGNERLLVYEHM 622
L DG +IAVK++ + ++G +F E +LT+V+HR++V+L GYC +G+E+LLVYE++
Sbjct: 300 LNDGREIAVKKLSR--RSNQGRTQFINEAKLLTRVQHRNVVSLFGYCAHGSEKLLVYEYV 473
Query: 623 PQGTLTQHLFECREHGYTPLTWKQRLIIALDVGRGVEYLHSLAQQSFIHRDLKPSNILLG 682
P+ +L + LF R L WK+R I V RG+ YLH + IHRD+K +NILL
Sbjct: 474 PRESLDKLLF--RSQKKEQLDWKRRFDIISGVARGLLYLHEDSHDCIIHRDIKAANILLD 647
Query: 683 DDMRAKVADFGLVKNAPDGNYSVET 707
+ K+ADFGL + P+ V T
Sbjct: 648 EKWVPKIADFGLARIFPEDQTHVNT 722
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.316 0.134 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,617,806
Number of Sequences: 28460
Number of extensions: 224328
Number of successful extensions: 2231
Number of sequences better than 10.0: 394
Number of HSP's better than 10.0 without gapping: 1711
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1969
length of query: 891
length of database: 4,897,600
effective HSP length: 98
effective length of query: 793
effective length of database: 2,108,520
effective search space: 1672056360
effective search space used: 1672056360
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 59 (27.3 bits)
Medicago: description of AC146585.14


