

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146574.14 - phase: 0
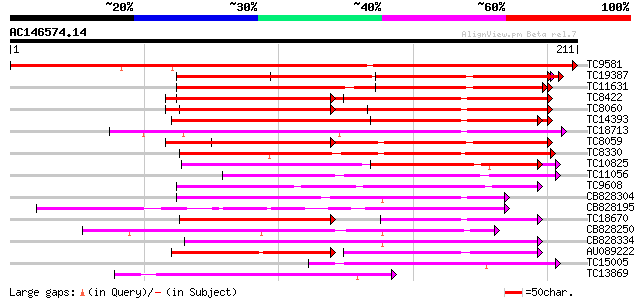
(211 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC9581 UP|Q9SCA1 (Q9SCA1) Calcium-binding protein, complete 321 7e-89
TC19387 weakly similar to UP|Q9FYK2 (Q9FYK2) F21J9.28, partial (... 109 3e-25
TC11631 weakly similar to UP|TCH2_ARATH (P25070) Calmodulin-rela... 109 4e-25
TC8422 UP|AAQ63461 (AAQ63461) Calmodulin 4 (Fragment), complete 108 9e-25
TC8060 UP|O49184 (O49184) Calmodulin (ESTS AU081349) (Auxin-regu... 107 2e-24
TC14393 similar to UP|Q93YA8 (Q93YA8) Calcium binding protein, p... 103 3e-23
TC18713 similar to GB|AAL90989.1|19699246|AY090328 AT3g03000/F13... 96 4e-21
TC8059 GB|AAB68399.1|1773321|HAU79736 calmodulin {Helianthus ann... 94 1e-20
TC8330 weakly similar to GB|AAM61257.1|21536925|AY084696 calmodu... 80 2e-16
TC10825 similar to GB|AAP68339.1|31711966|BT008900 At1g74740 {Ar... 75 6e-15
TC11056 similar to UP|O81390 (O81390) Calcium-dependent protein ... 74 1e-14
TC9608 similar to UP|Q94AZ4 (Q94AZ4) At1g12310/F5O11_2, partial ... 64 3e-11
CB828304 63 3e-11
CB828195 62 7e-11
TC18670 similar to UP|Q93YA8 (Q93YA8) Calcium binding protein, p... 60 4e-10
CB828250 57 3e-09
CB828334 56 4e-09
AU089222 53 3e-08
TC15005 homologue to UP|Q9AR92 (Q9AR92) Protein kinase , partia... 53 3e-08
TC13869 weakly similar to UP|Q9FR00 (Q9FR00) Avr9/Cf-9 rapidly e... 53 4e-08
>TC9581 UP|Q9SCA1 (Q9SCA1) Calcium-binding protein, complete
Length = 1049
Score = 321 bits (822), Expect = 7e-89
Identities = 168/232 (72%), Positives = 188/232 (80%), Gaps = 21/232 (9%)
Frame = +3
Query: 1 MPTILLRIFLLYNVVNSFLISLVPKKLITFFPHSWFTHQT----LTTPSSTSKRGLVFTK 56
MPTIL RIFLLYN++NSFL+SLVPKK+I F P SWF HQT ++ SS+S+ LV K
Sbjct: 96 MPTILHRIFLLYNLLNSFLLSLVPKKVIAFLPQSWFPHQTPSFSSSSSSSSSRGNLVIQK 275
Query: 57 TIT-----------------MDPNELKRVFQMFDRNDDGRITKKELNDSLENLGIFIPDK 99
T MDP ELKRVFQMFDRN DGRITKKELNDSLENLGIFIPDK
Sbjct: 276 TTDDCDPCQLLPLDTSLIPKMDPTELKRVFQMFDRNGDGRITKKELNDSLENLGIFIPDK 455
Query: 100 ELSQMIEKIDVNRDGCVDIEEFRELYESIMSERDEEEEEDMREAFNVFDQNGDGFISVDE 159
EL+QMIE+IDVN DGCVDI+EF ELY+SIM ERD EEEDMREAFNVFDQNGDGFI+V+E
Sbjct: 456 ELTQMIERIDVNGDGCVDIDEFGELYQSIMDERD--EEEDMREAFNVFDQNGDGFITVEE 629
Query: 160 LRSVLVSLGLKQGRTVEDCKKMIGTVDVDGNGLVDYKEFKQMMKGGGFTALS 211
LR+VL SLG+KQGRTVEDCKKMI VDVDG+G+VDYKEFKQMMKGGGF+AL+
Sbjct: 630 LRTVLASLGIKQGRTVEDCKKMIMKVDVDGDGMVDYKEFKQMMKGGGFSALT 785
>TC19387 weakly similar to UP|Q9FYK2 (Q9FYK2) F21J9.28, partial (45%)
Length = 631
Score = 109 bits (273), Expect = 3e-25
Identities = 59/140 (42%), Positives = 91/140 (64%)
Frame = +1
Query: 63 NELKRVFQMFDRNDDGRITKKELNDSLENLGIFIPDKELSQMIEKIDVNRDGCVDIEEFR 122
NE++ +F+ FDRN DG+I+ EL D + LG +E+ +M+ ++D N DG +D++EF
Sbjct: 43 NEVREIFKKFDRNGDGKISCSELKDLMAALGSKTTAEEVRRMMAELDRNGDGHIDLKEFG 222
Query: 123 ELYESIMSERDEEEEEDMREAFNVFDQNGDGFISVDELRSVLVSLGLKQGRTVEDCKKMI 182
E + +E +REAF ++D + +G IS EL SV+ LG K ++ DC+KMI
Sbjct: 223 EFHRGGGGGNSKE----IREAFEMYDLDKNGLISAKELHSVMKKLGEK--CSLGDCRKMI 384
Query: 183 GTVDVDGNGLVDYKEFKQMM 202
G VDVDG+G V+++EFK+MM
Sbjct: 385 GNVDVDGDGNVNFEEFKKMM 444
Score = 62.4 bits (150), Expect = 6e-11
Identities = 29/70 (41%), Positives = 47/70 (66%)
Frame = +1
Query: 137 EEDMREAFNVFDQNGDGFISVDELRSVLVSLGLKQGRTVEDCKKMIGTVDVDGNGLVDYK 196
+ ++RE F FD+NGDG IS EL+ ++ +LG K T E+ ++M+ +D +G+G +D K
Sbjct: 40 DNEVREIFKKFDRNGDGKISCSELKDLMAALGSK--TTAEEVRRMMAELDRNGDGHIDLK 213
Query: 197 EFKQMMKGGG 206
EF + +GGG
Sbjct: 214 EFGEFHRGGG 243
Score = 49.3 bits (116), Expect = 5e-07
Identities = 31/106 (29%), Positives = 51/106 (47%)
Frame = +1
Query: 98 DKELSQMIEKIDVNRDGCVDIEEFRELYESIMSERDEEEEEDMREAFNVFDQNGDGFISV 157
D E+ ++ +K D N DG + E ++L ++ S+ EE +R D+NGDG I +
Sbjct: 40 DNEVREIFKKFDRNGDGKISCSELKDLMAALGSKTTAEE---VRRMMAELDRNGDGHIDL 210
Query: 158 DELRSVLVSLGLKQGRTVEDCKKMIGTVDVDGNGLVDYKEFKQMMK 203
E G + + + +M D+D NGL+ KE +MK
Sbjct: 211 KEFGEFHRGGGGGNSKEIREAFEMY---DLDKNGLISAKELHSVMK 339
>TC11631 weakly similar to UP|TCH2_ARATH (P25070) Calmodulin-related protein
2, touch-induced, partial (68%)
Length = 530
Score = 109 bits (272), Expect = 4e-25
Identities = 55/140 (39%), Positives = 95/140 (67%)
Frame = +3
Query: 63 NELKRVFQMFDRNDDGRITKKELNDSLENLGIFIPDKELSQMIEKIDVNRDGCVDIEEFR 122
+E++++F FD+N DG+I+ EL + L LG +E+ +M+ +ID N DG +D++EF
Sbjct: 96 DEVRKIFNKFDKNGDGKISCTELKEMLCALGTKTSSEEVKRMMAEIDQNGDGYIDLKEFA 275
Query: 123 ELYESIMSERDEEEEEDMREAFNVFDQNGDGFISVDELRSVLVSLGLKQGRTVEDCKKMI 182
E + + + E++ +++R+A +++D + +G IS +EL SVL LG K ++ DC+KMI
Sbjct: 276 EFH---LHDAGEDDSKELRDACDLYDLDKNGLISANELHSVLKKLGEK--CSLGDCRKMI 440
Query: 183 GTVDVDGNGLVDYKEFKQMM 202
VD DG+G V+++EFK+MM
Sbjct: 441 SNVDADGDGNVNFEEFKKMM 500
Score = 58.5 bits (140), Expect = 8e-10
Identities = 27/64 (42%), Positives = 45/64 (70%)
Frame = +3
Query: 137 EEDMREAFNVFDQNGDGFISVDELRSVLVSLGLKQGRTVEDCKKMIGTVDVDGNGLVDYK 196
++++R+ FN FD+NGDG IS EL+ +L +LG K + E+ K+M+ +D +G+G +D K
Sbjct: 93 DDEVRKIFNKFDKNGDGKISCTELKEMLCALGTK--TSSEEVKRMMAEIDQNGDGYIDLK 266
Query: 197 EFKQ 200
EF +
Sbjct: 267 EFAE 278
>TC8422 UP|AAQ63461 (AAQ63461) Calmodulin 4 (Fragment), complete
Length = 832
Score = 108 bits (269), Expect = 9e-25
Identities = 59/140 (42%), Positives = 87/140 (62%)
Frame = +1
Query: 63 NELKRVFQMFDRNDDGRITKKELNDSLENLGIFIPDKELSQMIEKIDVNRDGCVDIEEFR 122
+E K F +FD++ DG IT KEL + +LG + EL MI ++D + +G +D EF
Sbjct: 166 SEFKEAFSLFDKDGDGSITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFL 345
Query: 123 ELYESIMSERDEEEEEDMREAFNVFDQNGDGFISVDELRSVLVSLGLKQGRTVEDCKKMI 182
L M + D EEE ++EAF VFD++ +GFIS ELR V+ +LG K T E+ +MI
Sbjct: 346 NLMARKMKDTDSEEE--LKEAFRVFDKDQNGFISAAELRHVMTNLGEK--LTDEEVDEMI 513
Query: 183 GTVDVDGNGLVDYKEFKQMM 202
DVDG+G ++Y+EF ++M
Sbjct: 514 READVDGDGQINYEEFVKVM 573
Score = 60.5 bits (145), Expect = 2e-10
Identities = 29/78 (37%), Positives = 50/78 (63%)
Frame = +1
Query: 125 YESIMSERDEEEEEDMREAFNVFDQNGDGFISVDELRSVLVSLGLKQGRTVEDCKKMIGT 184
+ ++ + +++ + +EAF++FD++GDG I+ EL +V+ SLG Q T + + MI
Sbjct: 127 HTTMADQLTDDQISEFKEAFSLFDKDGDGSITTKELGTVMRSLG--QNPTEAELQDMINE 300
Query: 185 VDVDGNGLVDYKEFKQMM 202
VD DGNG +D+ EF +M
Sbjct: 301 VDADGNGTIDFPEFLNLM 354
Score = 52.4 bits (124), Expect = 6e-08
Identities = 25/63 (39%), Positives = 40/63 (62%)
Frame = +1
Query: 59 TMDPNELKRVFQMFDRNDDGRITKKELNDSLENLGIFIPDKELSQMIEKIDVNRDGCVDI 118
T ELK F++FD++ +G I+ EL + NLG + D+E+ +MI + DV+ DG ++
Sbjct: 373 TDSEEELKEAFRVFDKDQNGFISAAELRHVMTNLGEKLTDEEVDEMIREADVDGDGQINY 552
Query: 119 EEF 121
EEF
Sbjct: 553 EEF 561
>TC8060 UP|O49184 (O49184) Calmodulin (ESTS AU081349) (Auxin-regulated
calmodulin) (Calmodulin TACAM1-1) (Calmodulin TACAM1-2)
(Calmodulin TACAM1-3) (Calmodulin TACAM3-1) (Calmodulin
TACAM3-2) (Calmodulin TACAM3-3) (Calmodulin TACAM4-1)
(CaM protein), complete
Length = 615
Score = 107 bits (266), Expect = 2e-24
Identities = 59/139 (42%), Positives = 86/139 (61%)
Frame = +3
Query: 64 ELKRVFQMFDRNDDGRITKKELNDSLENLGIFIPDKELSQMIEKIDVNRDGCVDIEEFRE 123
E K F +FD++ DG IT KEL + +LG + EL MI ++D + +G +D EF
Sbjct: 102 EFKEAFSLFDKDGDGCITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLN 281
Query: 124 LYESIMSERDEEEEEDMREAFNVFDQNGDGFISVDELRSVLVSLGLKQGRTVEDCKKMIG 183
L M + D EEE ++EAF VFD++ +GFIS ELR V+ +LG K T E+ +MI
Sbjct: 282 LMARKMKDTDSEEE--LKEAFRVFDKDQNGFISAAELRHVMTNLGEK--LTDEEVDEMIR 449
Query: 184 TVDVDGNGLVDYKEFKQMM 202
DVDG+G ++Y+EF ++M
Sbjct: 450 EADVDGDGQINYEEFVKVM 506
Score = 59.7 bits (143), Expect = 4e-10
Identities = 29/69 (42%), Positives = 46/69 (66%)
Frame = +3
Query: 134 EEEEEDMREAFNVFDQNGDGFISVDELRSVLVSLGLKQGRTVEDCKKMIGTVDVDGNGLV 193
+++ + +EAF++FD++GDG I+ EL +V+ SLG Q T + + MI VD DGNG +
Sbjct: 87 DDQIAEFKEAFSLFDKDGDGCITTKELGTVMRSLG--QNPTEAELQDMINEVDADGNGTI 260
Query: 194 DYKEFKQMM 202
D+ EF +M
Sbjct: 261 DFPEFLNLM 287
Score = 52.4 bits (124), Expect = 6e-08
Identities = 25/63 (39%), Positives = 40/63 (62%)
Frame = +3
Query: 59 TMDPNELKRVFQMFDRNDDGRITKKELNDSLENLGIFIPDKELSQMIEKIDVNRDGCVDI 118
T ELK F++FD++ +G I+ EL + NLG + D+E+ +MI + DV+ DG ++
Sbjct: 306 TDSEEELKEAFRVFDKDQNGFISAAELRHVMTNLGEKLTDEEVDEMIREADVDGDGQINY 485
Query: 119 EEF 121
EEF
Sbjct: 486 EEF 494
>TC14393 similar to UP|Q93YA8 (Q93YA8) Calcium binding protein, partial
(85%)
Length = 786
Score = 103 bits (256), Expect = 3e-23
Identities = 57/142 (40%), Positives = 88/142 (61%)
Frame = +3
Query: 61 DPNELKRVFQMFDRNDDGRITKKELNDSLENLGIFIPDKELSQMIEKIDVNRDGCVDIEE 120
D ELKRVF FD N DG+I+ EL++ L +LG +P +EL +++ +D + DG +++ E
Sbjct: 141 DMEELKRVFTRFDTNGDGKISVNELDNVLRSLGSGVPPEELKRVMVDLDGDHDGFINLSE 320
Query: 121 FRELYESIMSERDEEEEEDMREAFNVFDQNGDGFISVDELRSVLVSLGLKQGRTVEDCKK 180
F S ++ E + +AF ++DQ+ +G IS EL VL LG+ E+C K
Sbjct: 321 FAAFCRSDTADGGASE---LHDAFELYDQDKNGLISAAELCKVLNRLGMNCSE--EECHK 485
Query: 181 MIGTVDVDGNGLVDYKEFKQMM 202
MI +VD DG+G V+++EFK+MM
Sbjct: 486 MIKSVDSDGDGNVNFEEFKKMM 551
Score = 48.9 bits (115), Expect = 6e-07
Identities = 26/64 (40%), Positives = 41/64 (63%)
Frame = +3
Query: 135 EEEEDMREAFNVFDQNGDGFISVDELRSVLVSLGLKQGRTVEDCKKMIGTVDVDGNGLVD 194
++ E+++ F FD NGDG ISV+EL +VL SLG G E+ K+++ +D D +G ++
Sbjct: 138 DDMEELKRVFTRFDTNGDGKISVNELDNVLRSLG--SGVPPEELKRVMVDLDGDHDGFIN 311
Query: 195 YKEF 198
EF
Sbjct: 312 LSEF 323
>TC18713 similar to GB|AAL90989.1|19699246|AY090328 AT3g03000/F13E7_5
{Arabidopsis thaliana;}, partial (88%)
Length = 868
Score = 96.3 bits (238), Expect = 4e-21
Identities = 56/182 (30%), Positives = 95/182 (51%), Gaps = 12/182 (6%)
Frame = +1
Query: 38 HQTLTTPSSTS--------KRGLVFTKTITMDPN---ELKRVFQMFDRNDDGRITKKELN 86
HQ PSS+S K + + + +D EL+ +F+ FDRN+DG +T+ EL+
Sbjct: 103 HQIAIKPSSSSSLIPKKNHKEIMSKKQAVNLDEEQIAELREIFRSFDRNNDGSLTQLELS 282
Query: 87 DSLENLGIFIPDKELSQMIEKIDVNRDGCVDIEEF-RELYESIMSERDEEEEEDMREAFN 145
L +LG+ +++ I+K D N +G ++ EF + ++ R EE +R+ F
Sbjct: 283 SLLRSLGLKPSAEQVEGFIQKADTNSNGLIEFSEFVAVVAPELLPARSPYTEEQLRQLFR 462
Query: 146 VFDQNGDGFISVDELRSVLVSLGLKQGRTVEDCKKMIGTVDVDGNGLVDYKEFKQMMKGG 205
VFD++G+GFI+ EL + LG + E+ MI D DG+G++ ++EF +
Sbjct: 463 VFDRDGNGFITAAELAHSMAKLG--HALSAEELTGMIKEADADGDGMISFQEFAHAISSA 636
Query: 206 GF 207
F
Sbjct: 637 AF 642
>TC8059 GB|AAB68399.1|1773321|HAU79736 calmodulin {Helianthus annuus;} ,
partial (83%)
Length = 836
Score = 94.4 bits (233), Expect = 1e-20
Identities = 54/127 (42%), Positives = 78/127 (60%)
Frame = +2
Query: 76 DDGRITKKELNDSLENLGIFIPDKELSQMIEKIDVNRDGCVDIEEFRELYESIMSERDEE 135
D G IT KEL + +LG + EL MI ++D + +G +D EF L M + D E
Sbjct: 158 DPGCITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLNLMARKMKDTDSE 337
Query: 136 EEEDMREAFNVFDQNGDGFISVDELRSVLVSLGLKQGRTVEDCKKMIGTVDVDGNGLVDY 195
EE ++EAF VFD++ +GFIS ELR V+ +LG K T E+ +MI DVDG+G ++Y
Sbjct: 338 EE--LKEAFRVFDKDQNGFISAAELRHVMTNLGEK--LTDEEVDEMIREADVDGDGQINY 505
Query: 196 KEFKQMM 202
+EF ++M
Sbjct: 506 EEFVKVM 526
Score = 52.4 bits (124), Expect = 6e-08
Identities = 25/63 (39%), Positives = 40/63 (62%)
Frame = +2
Query: 59 TMDPNELKRVFQMFDRNDDGRITKKELNDSLENLGIFIPDKELSQMIEKIDVNRDGCVDI 118
T ELK F++FD++ +G I+ EL + NLG + D+E+ +MI + DV+ DG ++
Sbjct: 326 TDSEEELKEAFRVFDKDQNGFISAAELRHVMTNLGEKLTDEEVDEMIREADVDGDGQINY 505
Query: 119 EEF 121
EEF
Sbjct: 506 EEF 514
>TC8330 weakly similar to GB|AAM61257.1|21536925|AY084696 calmodulin-like
protein {Arabidopsis thaliana;} , partial (72%)
Length = 1174
Score = 80.5 bits (197), Expect = 2e-16
Identities = 47/141 (33%), Positives = 86/141 (60%), Gaps = 1/141 (0%)
Frame = +3
Query: 64 ELKRVFQMFDRNDDGRITKKELNDSLENLGIF-IPDKELSQMIEKIDVNRDGCVDIEEFR 122
+++ F++ DR++DG +++ +L L LG + D+E+ M+ ++D + G + +E
Sbjct: 291 DVRWAFRLIDRDNDGVVSRADLVAVLTRLGALQLSDEEVEVMLREVDGDGRGSISVEA-- 464
Query: 123 ELYESIMSERDEEEEEDMREAFNVFDQNGDGFISVDELRSVLVSLGLKQGRTVEDCKKMI 182
L E + D E E +REAF VFD + DG IS +EL V ++G + T+++C++MI
Sbjct: 465 -LMERVGPGSDPETE--LREAFEVFDTDRDGRISAEELLRVFTAIG-DERCTLDECRRMI 632
Query: 183 GTVDVDGNGLVDYKEFKQMMK 203
VD +G+G V +++F +MM+
Sbjct: 633 EGVDRNGDGFVCFEDFSRMME 695
>TC10825 similar to GB|AAP68339.1|31711966|BT008900 At1g74740 {Arabidopsis
thaliana;}, partial (32%)
Length = 1091
Score = 75.5 bits (184), Expect = 6e-15
Identities = 45/142 (31%), Positives = 75/142 (52%), Gaps = 1/142 (0%)
Frame = +1
Query: 65 LKRVFQMFDRNDDGRITKKELNDSLENLGIFIPDKELSQMIEKIDVNRDGCVDIEEFREL 124
+K +F + D + DGR++ +EL L +G + D+E+ ++E DV+ +G +D EF +
Sbjct: 19 IKDMFTLMDTDKDGRVSYEELKAGLRKVGSQLADQEIKMLMEVADVDGNGVLDYGEFVAV 198
Query: 125 YESIMSERDEEEEEDMREAFNVFDQNGDGFISVDELRSVLVSLGLKQGRTVED-CKKMIG 183
+ E +E +AF FD++G G+I EL+ L + G T D ++
Sbjct: 199 TIHLQKM---ENDEHFHKAFKFFDKDGSGYIESGELQEALAD---ESGVTDADVLNDIMR 360
Query: 184 TVDVDGNGLVDYKEFKQMMKGG 205
VD D +G + Y+EF MMK G
Sbjct: 361 EVDTDKDGRISYEEFVAMMKAG 426
Score = 45.8 bits (107), Expect = 5e-06
Identities = 23/64 (35%), Positives = 40/64 (61%)
Frame = +1
Query: 135 EEEEDMREAFNVFDQNGDGFISVDELRSVLVSLGLKQGRTVEDCKKMIGTVDVDGNGLVD 194
EE E +++ F + D + DG +S +EL++ L +G + ++ K ++ DVDGNG++D
Sbjct: 4 EEVEIIKDMFTLMDTDKDGRVSYEELKAGLRKVGSQLAD--QEIKMLMEVADVDGNGVLD 177
Query: 195 YKEF 198
Y EF
Sbjct: 178 YGEF 189
>TC11056 similar to UP|O81390 (O81390) Calcium-dependent protein kinase ,
partial (22%)
Length = 675
Score = 74.3 bits (181), Expect = 1e-14
Identities = 40/126 (31%), Positives = 70/126 (54%)
Frame = +3
Query: 80 ITKKELNDSLENLGIFIPDKELSQMIEKIDVNRDGCVDIEEFRELYESIMSERDEEEEED 139
IT +EL L +G + + E+ Q++E DV+ +G +D EF + M E +E
Sbjct: 6 ITYEELKSGLARIGSRLSETEVKQLMEAADVDGNGSIDYLEF---ISATMHRHRLERDEH 176
Query: 140 MREAFNVFDQNGDGFISVDELRSVLVSLGLKQGRTVEDCKKMIGTVDVDGNGLVDYKEFK 199
+ +AF FD++ G I++DEL + + G+ ++ K++I VD D +G ++Y+EF
Sbjct: 177 LYKAFQYFDKDNSGHITIDELETAMTQHGMGDEASI---KEIISEVDTDNDGRINYEEFC 347
Query: 200 QMMKGG 205
MM+ G
Sbjct: 348 AMMRSG 365
>TC9608 similar to UP|Q94AZ4 (Q94AZ4) At1g12310/F5O11_2, partial (97%)
Length = 817
Score = 63.5 bits (153), Expect = 3e-11
Identities = 42/136 (30%), Positives = 68/136 (49%)
Frame = +3
Query: 63 NELKRVFQMFDRNDDGRITKKELNDSLENLGIFIPDKELSQMIEKIDVNRDGCVDIEEFR 122
+ +K F +FD + DG+I EL + +LG +L ++ + N D F
Sbjct: 135 SSMKEAFTLFDTDGDGKIAPSELGILMRSLGGNPTQAQLKSIVA--EENLTSPFDFPRFL 308
Query: 123 ELYESIMSERDEEEEEDMREAFNVFDQNGDGFISVDELRSVLVSLGLKQGRTVEDCKKMI 182
+L M + E + +R+AF V D++ GF+SV ELR +L S+G K D + I
Sbjct: 309 DLMAKHM--KPEPFDRQLRDAFKVIDKDSTGFVSVTELRHILTSIGEKLEPAEFD--EWI 476
Query: 183 GTVDVDGNGLVDYKEF 198
VDV +G + Y++F
Sbjct: 477 REVDVGSDGKIRYEDF 524
Score = 35.4 bits (80), Expect = 0.007
Identities = 18/54 (33%), Positives = 33/54 (60%), Gaps = 1/54 (1%)
Frame = +3
Query: 116 VDIEEFRELYESIMS-ERDEEEEEDMREAFNVFDQNGDGFISVDELRSVLVSLG 168
V + + + + E+ M + +++ M+EAF +FD +GDG I+ EL ++ SLG
Sbjct: 66 VALSQHKRIIETKMGKDLSDDQVSSMKEAFTLFDTDGDGKIAPSELGILMRSLG 227
>CB828304
Length = 489
Score = 63.2 bits (152), Expect = 3e-11
Identities = 40/129 (31%), Positives = 65/129 (50%), Gaps = 5/129 (3%)
Frame = +1
Query: 63 NELKRVFQMFDRNDDGRITKKELNDSLENLGIFIPDKELSQMIEKIDVNRDGCVDIEEFR 122
N+L+ +F FD + DG +T EL L +LG+ +L ++ +D N +G V EF
Sbjct: 115 NQLREIFARFDMDSDGSLTMLELAALLRSLGLKPSGDQLHDLLSNMDSNGNGSV---EFD 285
Query: 123 ELYESIMSERDEEEE-----EDMREAFNVFDQNGDGFISVDELRSVLVSLGLKQGRTVED 177
EL +I+ + E E + + F FD++ +GFIS EL + +G Q T ++
Sbjct: 286 ELVRTILPDLKNNAEVLLNQEQLLDVFKCFDRDSNGFISAAELAGAMAKMG--QPLTYKE 459
Query: 178 CKKMIGTVD 186
+MI D
Sbjct: 460 LTEMIREAD 486
Score = 35.4 bits (80), Expect = 0.007
Identities = 17/52 (32%), Positives = 32/52 (60%)
Frame = +1
Query: 58 ITMDPNELKRVFQMFDRNDDGRITKKELNDSLENLGIFIPDKELSQMIEKID 109
+ ++ +L VF+ FDR+ +G I+ EL ++ +G + KEL++MI + D
Sbjct: 331 VLLNQEQLLDVFKCFDRDSNGFISAAELAGAMAKMGQPLTYKELTEMIREAD 486
>CB828195
Length = 551
Score = 62.0 bits (149), Expect = 7e-11
Identities = 51/176 (28%), Positives = 78/176 (43%)
Frame = +3
Query: 11 LYNVVNSFLISLVPKKLITFFPHSWFTHQTLTTPSSTSKRGLVFTKTITMDPNELKRVFQ 70
L+ +++ FL + K++ FF + W ++ + S NE K
Sbjct: 111 LFGLIDLFLYCTIFNKILKFFSNFWCSNSEVRGEEKVSDY------EFNHQENESK---- 260
Query: 71 MFDRNDDGRITKKELNDSLENLGIFIPDKELSQMIEKIDVNRDGCVDIEEFRELYESIMS 130
+G I + E+ + LG F KE ++ EK +E EL+E
Sbjct: 261 -----SEG-IQRDEVKKVMAELGFFC-SKESEELEEKYGS--------KELSELFED--- 386
Query: 131 ERDEEEEEDMREAFNVFDQNGDGFISVDELRSVLVSLGLKQGRTVEDCKKMIGTVD 186
E E++++AF+VFD+N DGFI EL VL LGLK+ +E CK MI D
Sbjct: 387 --QEPSLEEVKQAFDVFDENRDGFIDARELHRVLCVLGLKEEAGIEKCKIMIRNFD 548
>TC18670 similar to UP|Q93YA8 (Q93YA8) Calcium binding protein, partial
(41%)
Length = 329
Score = 59.7 bits (143), Expect = 4e-10
Identities = 27/58 (46%), Positives = 39/58 (66%)
Frame = +3
Query: 64 ELKRVFQMFDRNDDGRITKKELNDSLENLGIFIPDKELSQMIEKIDVNRDGCVDIEEF 121
EL+RVF FD N DG+I+ EL+ L LG +P +EL Q++E +D +RDG + + EF
Sbjct: 114 ELERVFNRFDANSDGKISVDELDSVLRTLGSGVPPEELQQVMEDLDTDRDGFISLPEF 287
Score = 46.2 bits (108), Expect = 4e-06
Identities = 25/60 (41%), Positives = 36/60 (59%)
Frame = +3
Query: 139 DMREAFNVFDQNGDGFISVDELRSVLVSLGLKQGRTVEDCKKMIGTVDVDGNGLVDYKEF 198
++ FN FD N DG ISVDEL SVL +LG G E+ ++++ +D D +G + EF
Sbjct: 114 ELERVFNRFDANSDGKISVDELDSVLRTLG--SGVPPEELQQVMEDLDTDRDGFISLPEF 287
>CB828250
Length = 485
Score = 56.6 bits (135), Expect = 3e-09
Identities = 45/159 (28%), Positives = 77/159 (48%), Gaps = 4/159 (2%)
Frame = +1
Query: 28 ITFFPHSWFTHQTLTT-PSSTSKRGLVFTKTITMDPNELKRVFQMFDRNDDGRITKKELN 86
++ FP + T Q T PS S R + + ++ + F + D + DG+I++ +L
Sbjct: 4 VSVFPSTIKTTQHNTMCPSGRSLRSNASSSSSLSATSDFRPAFDVLDADCDGKISRDDLR 183
Query: 87 DSLENL--GIFIPDKELSQMIEKIDVNRDGCVDIEEFRELYESIMSERDEEEE-EDMREA 143
+ G+ D + M+ D ++DG V+ EEF E +++E ++ M E
Sbjct: 184 AFYAGIYGGVASGDDVIGAMMTVADTDKDGFVEYEEF----ERVVTENNKPLGCGAMEEV 351
Query: 144 FNVFDQNGDGFISVDELRSVLVSLGLKQGRTVEDCKKMI 182
F V D++GDG +S +L+S + S G T ED MI
Sbjct: 352 FKVMDKDGDGKLSHHDLKSYMASAGF--SATDEDINAMI 462
Score = 34.7 bits (78), Expect = 0.013
Identities = 23/102 (22%), Positives = 44/102 (42%)
Frame = +1
Query: 108 IDVNRDGCVDIEEFRELYESIMSERDEEEEEDMREAFNVFDQNGDGFISVDELRSVLVSL 167
+D + DG + ++ R Y I ++ + V D + DGF+ +E V+
Sbjct: 139 LDADCDGKISRDDLRAFYAGIYGGVASGDDV-IGAMMTVADTDKDGFVEYEEFERVVTEN 315
Query: 168 GLKQGRTVEDCKKMIGTVDVDGNGLVDYKEFKQMMKGGGFTA 209
G + ++ +D DG+G + + + K M GF+A
Sbjct: 316 NKPLGCGAME--EVFKVMDKDGDGKLSHHDLKSYMASAGFSA 435
>CB828334
Length = 541
Score = 56.2 bits (134), Expect = 4e-09
Identities = 36/149 (24%), Positives = 66/149 (44%), Gaps = 16/149 (10%)
Frame = +2
Query: 66 KRVFQMFDRNDDGRITKKELNDSLENLGIFIPDKELSQMIEKIDVNRDGCVDIEEFRELY 125
K +F+ FD + +G I ++EL L I ++E++ + E D+N D ++ EF L
Sbjct: 56 KAIFKQFDEDSNGAIDQEELKKCFSKLEISFTEEEVNDLFEACDINEDMGMEFSEFIVLL 235
Query: 126 ESIMSERDEEEE----------------EDMREAFNVFDQNGDGFISVDELRSVLVSLGL 169
+ +D+ E + + F D+N DG++S E+ +
Sbjct: 236 CLVHLLKDDRAAVQAKSRIGMPDLETTFETLVDTFVFLDKNKDGYVSKSEMVQAINETVS 415
Query: 170 KQGRTVEDCKKMIGTVDVDGNGLVDYKEF 198
+ + K +D D NG+V++KEF
Sbjct: 416 GERSSGRIAMKRFEEMDWDKNGMVNFKEF 502
>AU089222
Length = 345
Score = 53.1 bits (126), Expect = 3e-08
Identities = 28/74 (37%), Positives = 40/74 (53%)
Frame = +3
Query: 125 YESIMSERDEEEEEDMREAFNVFDQNGDGFISVDELRSVLVSLGLKQGRTVEDCKKMIGT 184
Y I D ++ D F FD NGDG IS EL L +LG T E+ ++M+
Sbjct: 33 YILIKMADDPQDVADRERIFKRFDANGDGQISSSELGEALKTLG---SVTAEEVQRMMDE 203
Query: 185 VDVDGNGLVDYKEF 198
+D+DG+G + Y+EF
Sbjct: 204 IDIDGDGFISYEEF 245
Score = 51.2 bits (121), Expect = 1e-07
Identities = 23/61 (37%), Positives = 42/61 (68%)
Frame = +3
Query: 61 DPNELKRVFQMFDRNDDGRITKKELNDSLENLGIFIPDKELSQMIEKIDVNRDGCVDIEE 120
D + +R+F+ FD N DG+I+ EL ++L+ LG + +E+ +M+++ID++ DG + EE
Sbjct: 66 DVADRERIFKRFDANGDGQISSSELGEALKTLG-SVTAEEVQRMMDEIDIDGDGFISYEE 242
Query: 121 F 121
F
Sbjct: 243 F 245
Score = 34.7 bits (78), Expect = 0.013
Identities = 22/67 (32%), Positives = 35/67 (51%)
Frame = +3
Query: 96 IPDKELSQMIEKIDVNRDGCVDIEEFRELYESIMSERDEEEEEDMREAFNVFDQNGDGFI 155
+ D+E ++ ++ D N DG + E E +++ S EE + M E D +GDGFI
Sbjct: 69 VADRE--RIFKRFDANGDGQISSSELGEALKTLGSVTAEEVQRMMDE----IDIDGDGFI 230
Query: 156 SVDELRS 162
S +E S
Sbjct: 231 SYEEFTS 251
>TC15005 homologue to UP|Q9AR92 (Q9AR92) Protein kinase , partial (19%)
Length = 657
Score = 53.1 bits (126), Expect = 3e-08
Identities = 31/98 (31%), Positives = 50/98 (50%), Gaps = 4/98 (4%)
Frame = +2
Query: 112 RDGCVDIEEFRELYESIMSERDEEEEEDMREAFNVFDQNGDGFISVDELRSVLVSLGLKQ 171
RDG +D EF + M E ++ + AF FD++ GFI+ DEL + G+
Sbjct: 11 RDGTIDYIEF---VTATMHRHKLERDDHLN*AFPYFDKDSSGFITRDELEIAMKEYGMGD 181
Query: 172 GRTVE----DCKKMIGTVDVDGNGLVDYKEFKQMMKGG 205
T++ + +I VD D +G ++Y+EF MM+ G
Sbjct: 182 DATIKEIISEVDTIISEVDTDHDGRINYEEFCAMMRSG 295
>TC13869 weakly similar to UP|Q9FR00 (Q9FR00) Avr9/Cf-9 rapidly elicited
protein 31, partial (45%)
Length = 547
Score = 52.8 bits (125), Expect = 4e-08
Identities = 34/111 (30%), Positives = 60/111 (53%), Gaps = 6/111 (5%)
Frame = +3
Query: 40 TLTTPSSTSKRGLVFTKTITMDPNELKRVFQMFDRNDDGRITKKELNDSLENLGIFIPDK 99
TLT+PS + + ++ +++ L+R+F MFD+N D IT +E++ +L LG+
Sbjct: 228 TLTSPSRSFR-----LRSPSLNSLRLRRIFDMFDKNGDCMITVEEISQALNLLGLEAEVA 392
Query: 100 ELSQMIEKIDVNRDGCVDIEEFRELYESI------MSERDEEEEEDMREAF 144
E+ MI + + ++F L+ESI E ++ +E D+REAF
Sbjct: 393 EIDSMIRSYIRPGNEGLTYDDFMALHESIGDTFFGFVEEEKGDESDLREAF 545
Score = 37.0 bits (84), Expect = 0.003
Identities = 23/69 (33%), Positives = 37/69 (53%)
Frame = +3
Query: 140 MREAFNVFDQNGDGFISVDELRSVLVSLGLKQGRTVEDCKKMIGTVDVDGNGLVDYKEFK 199
+R F++FD+NGD I+V+E+ L LGL+ V + MI + GN + Y +F
Sbjct: 288 LRRIFDMFDKNGDCMITVEEISQALNLLGLE--AEVAEIDSMIRSYIRPGNEGLTYDDFM 461
Query: 200 QMMKGGGFT 208
+ + G T
Sbjct: 462 ALHESIGDT 488
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.320 0.139 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,833,898
Number of Sequences: 28460
Number of extensions: 33118
Number of successful extensions: 406
Number of sequences better than 10.0: 73
Number of HSP's better than 10.0 without gapping: 337
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 368
length of query: 211
length of database: 4,897,600
effective HSP length: 86
effective length of query: 125
effective length of database: 2,450,040
effective search space: 306255000
effective search space used: 306255000
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 53 (25.0 bits)
Medicago: description of AC146574.14


