

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146573.11 + phase: 2 /pseudo
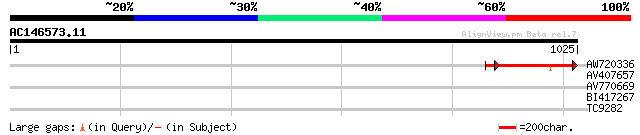
(1025 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
AW720336 183 1e-46
AV407657 35 0.079
AV770669 32 0.51
BI417267 29 4.3
TC9282 similar to UP|Q94LX7 (Q94LX7) Phosphoenolpyruvate carboxy... 28 9.7
>AW720336
Length = 559
Score = 183 bits (465), Expect = 1e-46
Identities = 93/155 (60%), Positives = 116/155 (74%), Gaps = 5/155 (3%)
Frame = +1
Query: 876 FLSEKIGKQRNGFEYAALEVKNYLERYMFSNIVSTVMIIFTPQHCSQVKYSPWVCHFHLD 935
F + + GFEYAALEVKNYLER MFS+IVSTVMI+FTPQ S ++PWVCHFHLD
Sbjct: 76 FCRTNLVNRGTGFEYAALEVKNYLERVMFSDIVSTVMIMFTPQSSSLEIFNPWVCHFHLD 255
Query: 936 KENVTRRKLEVHSIIDSLYQQYDSFRRESKVILPSLKISSN-----EGGETSVGNEKDGE 990
KE V RRKL VHS+I+SLY++Y+S +ESKV P+LKISSN EGG S+ EK+
Sbjct: 256 KEIVARRKLTVHSVIESLYRRYESLTKESKVTFPNLKISSNRKCSKEGGYASLNKEKEDV 435
Query: 991 DCITVAVVEDSKNSVQLDSVRKSLIPFLLQTAIKG 1025
DCI+V +VE S++S +L++VR +IPFLL T IKG
Sbjct: 436 DCISVTIVESSRSSAKLEAVRDLMIPFLLGTVIKG 540
Score = 51.2 bits (121), Expect = 8e-07
Identities = 23/26 (88%), Positives = 25/26 (95%)
Frame = +3
Query: 860 LECGSRKKGGDQTVSLFLSEKIGKQR 885
LECGSRKK GDQTVSLFLS+K+GKQR
Sbjct: 27 LECGSRKKSGDQTVSLFLSDKLGKQR 104
>AV407657
Length = 426
Score = 34.7 bits (78), Expect = 0.079
Identities = 17/26 (65%), Positives = 20/26 (76%)
Frame = +1
Query: 233 NQGLQEIG*FQGYSK*VKFSCFGLPE 258
+QGLQEIG*FQ S+*VK SC P+
Sbjct: 145 DQGLQEIG*FQRSSR*VKLSCHQFPK 222
>AV770669
Length = 469
Score = 32.0 bits (71), Expect = 0.51
Identities = 15/43 (34%), Positives = 23/43 (52%)
Frame = -2
Query: 107 RVKVCLLPIVKVLFHAYSVLNHSLDTIISPKVANIAVGSLSCH 149
R ++C LP++ L H Y + L T + P +I+ SLS H
Sbjct: 402 RYRICFLPLITFLEHEYGIAFLPLITFLPPITLHISHFSLSIH 274
>BI417267
Length = 628
Score = 28.9 bits (63), Expect = 4.3
Identities = 18/62 (29%), Positives = 28/62 (45%), Gaps = 9/62 (14%)
Frame = -2
Query: 479 FWSGKQLFSMLLPSKFDYAFPSNDVFVKDGELISCS---------ESSGWLRDSENNVFQ 529
+WS LL KF F S +VF+ + C+ S GWL + +NN+F
Sbjct: 267 YWSSGNDGQTLLWEKFQQLFSS*NVFLT--RYVRCNLTIF**LRMRSRGWLSNIDNNIFS 94
Query: 530 SL 531
++
Sbjct: 93 AI 88
>TC9282 similar to UP|Q94LX7 (Q94LX7) Phosphoenolpyruvate carboxykinase ,
partial (44%)
Length = 1236
Score = 27.7 bits (60), Expect = 9.7
Identities = 15/39 (38%), Positives = 20/39 (50%)
Frame = -2
Query: 5 QKSF*LTCNLLISYLCYLVLCLQEKISALEINAPGQVTC 43
+ SF + NL Y C+L LCLQE A +N +C
Sbjct: 713 RNSFHFSRNLKSKYRCFLELCLQE---ASRVNGINNFSC 606
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.328 0.141 0.432
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 18,172,013
Number of Sequences: 28460
Number of extensions: 260492
Number of successful extensions: 1649
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 1637
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1648
length of query: 1025
length of database: 4,897,600
effective HSP length: 99
effective length of query: 926
effective length of database: 2,080,060
effective search space: 1926135560
effective search space used: 1926135560
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 60 (27.7 bits)
Medicago: description of AC146573.11


