

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146571.4 + phase: 0
(494 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
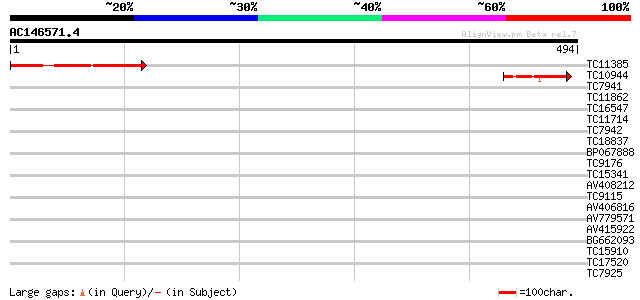
Score E
Sequences producing significant alignments: (bits) Value
TC11385 similar to UP|Q944Q7 (Q944Q7) At1g54990/F14C21_5, partia... 114 5e-26
TC10944 91 6e-19
TC7941 similar to UP|O49857 (O49857) Epoxide hydrolase , partia... 37 0.007
TC11862 similar to UP|Q39856 (Q39856) Epoxide hydrolase , parti... 36 0.013
TC16547 similar to UP|O49857 (O49857) Epoxide hydrolase , parti... 36 0.016
TC11714 similar to UP|Q16861 (Q16861) Super cysteine rich protei... 36 0.016
TC7942 similar to UP|Q8LPE6 (Q8LPE6) Epoxide hydrolase (Fragmen... 35 0.021
TC18837 weakly similar to UP|Q39856 (Q39856) Epoxide hydrolase ... 35 0.028
BP067888 35 0.028
TC9176 35 0.037
TC15341 similar to UP|Q9M0H8 (Q9M0H8) Predicted proline-rich pro... 34 0.048
AV408212 34 0.048
TC9115 similar to PIR|T00425|T00425 photolyase/blue-light recept... 32 0.18
AV406816 32 0.24
AV779571 32 0.31
AV415922 32 0.31
BG662093 31 0.40
TC15910 similar to UP|Q9AR64 (Q9AR64) Urease accessory protein G... 31 0.53
TC17520 similar to GB|AAO63388.1|28950929|BT005324 At5g19850 {Ar... 30 0.69
TC7925 homologue to UP|RPE_SPIOL (Q43157) Ribulose-phosphate 3-e... 30 1.2
>TC11385 similar to UP|Q944Q7 (Q944Q7) At1g54990/F14C21_5, partial (12%)
Length = 448
Score = 114 bits (284), Expect = 5e-26
Identities = 61/119 (51%), Positives = 77/119 (64%)
Frame = +3
Query: 1 MAIITEEQESDQQQQQQKQKQKQKQQHSNKTKSKTQKPSTTQPQQTNPLSFWFYFTLSIS 60
MAIITEE+E +++ Q K Q + S+ S++ T P SFWFYFTLS+S
Sbjct: 120 MAIITEEEEEEEKPHQLKHPQIPPTKPSS---------SSSTSNTTEPFSFWFYFTLSVS 272
Query: 61 LVTLFFLFTSSPLSSSSPQQWFLTLPTTLRQHYSNGRTIKVQIHPNQQPIHLFTFQLDP 119
L+TL F+FTSS LS + WFL+LP+TLRQHYS GR IKVQ PNQ PI +FT + P
Sbjct: 273 LITLTFVFTSS-LSPQDSKAWFLSLPSTLRQHYSQGRIIKVQTRPNQPPIEVFTVEDGP 446
>TC10944
Length = 512
Score = 90.5 bits (223), Expect = 6e-19
Identities = 44/63 (69%), Positives = 50/63 (78%), Gaps = 4/63 (6%)
Frame = +1
Query: 431 TVRKVEQEPPIPDHVQKMFDEAKSDDGHDH----HQGHSHGHDRLGEAHIHEAGYMDAYG 486
TVRKVEQEP IPDH+Q+MFDEAKS+ GHDH H H HG+D G+AH H AGYMDAYG
Sbjct: 1 TVRKVEQEP-IPDHIQEMFDEAKSN-GHDHDHLHHHSHGHGYDHHGDAHDHGAGYMDAYG 174
Query: 487 IGH 489
+GH
Sbjct: 175 LGH 183
>TC7941 similar to UP|O49857 (O49857) Epoxide hydrolase , partial (48%)
Length = 610
Score = 37.0 bits (84), Expect = 0.007
Identities = 25/88 (28%), Positives = 42/88 (47%)
Frame = +2
Query: 86 PTTLRQHYSNGRTIKVQIHPNQQPIHLFTFQLDPTIPNPSETVLILHGQALSSYSYRNLI 145
P RQ + R ++ +H + + + ++ VL LHG YS+R+ I
Sbjct: 74 PVAARQTENENREMESIVH---RTVEVNGIKMHVAEKGEGPVVLFLHGFPELWYSWRHQI 244
Query: 146 QSLSTQGVRVIAIDLPGSGFSDKSVEVS 173
SLS++G R +A DL G G ++ V+
Sbjct: 245 LSLSSRGYRAVAPDLRGYGDTEAPSSVT 328
>TC11862 similar to UP|Q39856 (Q39856) Epoxide hydrolase , partial (50%)
Length = 564
Score = 36.2 bits (82), Expect = 0.013
Identities = 22/55 (40%), Positives = 30/55 (54%)
Frame = +1
Query: 128 VLILHGQALSSYSYRNLIQSLSTQGVRVIAIDLPGSGFSDKSVEVSVEGLDGIFG 182
VL LHG YS+R+ I +LS+QG R +A DL G G ++ V+ I G
Sbjct: 85 VLFLHGFPELWYSWRHQILALSSQGYRAVAPDLRGYGDTEAPPSVTSYNCFNIVG 249
>TC16547 similar to UP|O49857 (O49857) Epoxide hydrolase , partial (50%)
Length = 550
Score = 35.8 bits (81), Expect = 0.016
Identities = 18/46 (39%), Positives = 28/46 (60%)
Frame = +1
Query: 128 VLILHGQALSSYSYRNLIQSLSTQGVRVIAIDLPGSGFSDKSVEVS 173
VL LHG YS+R+ I +LS++G +A DL G G +D + ++
Sbjct: 121 VLFLHGFPELWYSWRHQILTLSSRGYHAVAPDLRGYGDTDSPISIT 258
>TC11714 similar to UP|Q16861 (Q16861) Super cysteine rich protein
(Fragment), partial (48%)
Length = 519
Score = 35.8 bits (81), Expect = 0.016
Identities = 17/44 (38%), Positives = 27/44 (60%)
Frame = -1
Query: 7 EQESDQQQQQQKQKQKQKQQHSNKTKSKTQKPSTTQPQQTNPLS 50
+++ QQQQ Q+ KQ+Q+Q KT++ Q P Q QQT ++
Sbjct: 138 KKQQQQQQQLQQLKQQQQQLQQQKTRNSKQVPRH*QQQQTTTVT 7
>TC7942 similar to UP|Q8LPE6 (Q8LPE6) Epoxide hydrolase (Fragment) ,
partial (91%)
Length = 1288
Score = 35.4 bits (80), Expect = 0.021
Identities = 20/46 (43%), Positives = 28/46 (60%)
Frame = +3
Query: 128 VLILHGQALSSYSYRNLIQSLSTQGVRVIAIDLPGSGFSDKSVEVS 173
VL LHG YS+R+ I SLS++G R +A DL G G ++ V+
Sbjct: 75 VLFLHGFPELWYSWRHQILSLSSRGYRAVAPDLRGYGDTEAPSSVT 212
>TC18837 weakly similar to UP|Q39856 (Q39856) Epoxide hydrolase , partial
(89%)
Length = 1100
Score = 35.0 bits (79), Expect = 0.028
Identities = 20/46 (43%), Positives = 26/46 (56%)
Frame = +1
Query: 128 VLILHGQALSSYSYRNLIQSLSTQGVRVIAIDLPGSGFSDKSVEVS 173
+L LHG YS+R+ + SLS G R IA DL G G +D + S
Sbjct: 70 ILFLHGFPELWYSWRHQLLSLSALGYRAIAPDLRGYGDTDAPTDSS 207
>BP067888
Length = 534
Score = 35.0 bits (79), Expect = 0.028
Identities = 20/46 (43%), Positives = 26/46 (56%)
Frame = +3
Query: 128 VLILHGQALSSYSYRNLIQSLSTQGVRVIAIDLPGSGFSDKSVEVS 173
VL LHG Y++R+ I SLS G R +A DL G G +D V+
Sbjct: 132 VLFLHGFPELWYTWRHQILSLSALGYRAVAPDLRGYGDTDAPASVT 269
>TC9176
Length = 671
Score = 34.7 bits (78), Expect = 0.037
Identities = 18/40 (45%), Positives = 26/40 (65%)
Frame = +1
Query: 128 VLILHGQALSSYSYRNLIQSLSTQGVRVIAIDLPGSGFSD 167
VL +HG SYS+R+ I +L++ G R +A DL G G +D
Sbjct: 130 VLFIHGFPDLSYSWRHQITALASLGYRCVAPDLRGYGDTD 249
>TC15341 similar to UP|Q9M0H8 (Q9M0H8) Predicted proline-rich protein,
partial (33%)
Length = 1002
Score = 34.3 bits (77), Expect = 0.048
Identities = 18/44 (40%), Positives = 24/44 (53%)
Frame = +1
Query: 7 EQESDQQQQQQKQKQKQKQQHSNKTKSKTQKPSTTQPQQTNPLS 50
+ + QQQQQQ+Q+Q+Q Q + TQ PS PQ P S
Sbjct: 850 QYQQPQQQQQQQQQQQQWPQQLPQQVQPTQPPSMQSPQIRPPSS 981
>AV408212
Length = 362
Score = 34.3 bits (77), Expect = 0.048
Identities = 14/33 (42%), Positives = 25/33 (75%)
Frame = +3
Query: 2 AIITEEQESDQQQQQQKQKQKQKQQHSNKTKSK 34
A+ ++Q+ QQQQQQ+Q+Q Q+QQ ++ +S+
Sbjct: 84 ALQQQQQQQQQQQQQQQQQQSQQQQPQSQQQSR 182
Score = 33.5 bits (75), Expect = 0.081
Identities = 15/36 (41%), Positives = 25/36 (68%)
Frame = +3
Query: 12 QQQQQQKQKQKQKQQHSNKTKSKTQKPSTTQPQQTN 47
QQQQQQ+Q+Q+Q+QQ S + + ++Q+ S + N
Sbjct: 96 QQQQQQQQQQQQQQQQSQQQQPQSQQQSRDRAHLLN 203
Score = 32.7 bits (73), Expect = 0.14
Identities = 12/31 (38%), Positives = 24/31 (76%)
Frame = +3
Query: 6 EEQESDQQQQQQKQKQKQKQQHSNKTKSKTQ 36
++Q+ QQQQQQ+Q+Q+Q QQ +++ +++
Sbjct: 90 QQQQQQQQQQQQQQQQQQSQQQQPQSQQQSR 182
Score = 32.0 bits (71), Expect = 0.24
Identities = 16/40 (40%), Positives = 28/40 (70%)
Frame = +3
Query: 3 IITEEQESDQQQQQQKQKQKQKQQHSNKTKSKTQKPSTTQ 42
I+ + + QQQQQQ+Q+Q+Q+QQ + +S+ Q+P + Q
Sbjct: 63 ILLQRHAALQQQQQQQQQQQQQQQ---QQQSQQQQPQSQQ 173
Score = 31.2 bits (69), Expect = 0.40
Identities = 12/25 (48%), Positives = 20/25 (80%)
Frame = +3
Query: 2 AIITEEQESDQQQQQQKQKQKQKQQ 26
A + ++Q+ QQQQQQ+Q+Q+ +QQ
Sbjct: 81 AALQQQQQQQQQQQQQQQQQQSQQQ 155
Score = 30.8 bits (68), Expect = 0.53
Identities = 11/29 (37%), Positives = 21/29 (71%)
Frame = +3
Query: 6 EEQESDQQQQQQKQKQKQKQQHSNKTKSK 34
++Q+ QQQQQQ+Q Q+Q+ Q +++ +
Sbjct: 102 QQQQQQQQQQQQQQSQQQQPQSQQQSRDR 188
Score = 28.9 bits (63), Expect = 2.0
Identities = 10/28 (35%), Positives = 20/28 (70%)
Frame = +3
Query: 6 EEQESDQQQQQQKQKQKQKQQHSNKTKS 33
++Q+ QQQQQQ Q+Q+ + Q ++ ++
Sbjct: 108 QQQQQQQQQQQQSQQQQPQSQQQSRDRA 191
Score = 28.1 bits (61), Expect = 3.4
Identities = 17/53 (32%), Positives = 24/53 (45%), Gaps = 9/53 (16%)
Frame = +3
Query: 7 EQESDQQQQQQKQKQKQK---------QQHSNKTKSKTQKPSTTQPQQTNPLS 50
+Q QQQQQQ+ Q Q+ QQ + + + Q+ Q QQ P S
Sbjct: 9 QQSPHQQQQQQQHMQMQQILLQRHAALQQQQQQQQQQQQQQQQQQSQQQQPQS 167
>TC9115 similar to PIR|T00425|T00425 photolyase/blue-light receptor (PHR2)
[imported] - Arabidopsis thaliana
{Arabidopsis thaliana;}, partial (55%)
Length = 1018
Score = 32.3 bits (72), Expect = 0.18
Identities = 35/119 (29%), Positives = 47/119 (39%), Gaps = 21/119 (17%)
Frame = +2
Query: 27 HSNKTKSKTQKPSTT----QPQQ----TNPL-------SFWFYFTLS------ISLVTLF 65
HS K+K+ Q PSTT + QQ T P+ S FTL + +F
Sbjct: 44 HSPKSKNGVQFPSTTTIIRERQQR*PETTPIRTITIRSSITLPFTLHHPPLRPTQIPIIF 223
Query: 66 FLFTSSPLSSSSPQQWFLTLPTTLRQHYSNGRTIKVQIHPNQQPIHLFTFQLDPTIPNP 124
F T++ S P TL H + I +QIHP +QP + P P P
Sbjct: 224 FPTTTTQQGQSPNPSLHPHPPLTLHHHLTLAAQILLQIHPLRQPPPRPSLLRPPPPPRP 400
>AV406816
Length = 417
Score = 32.0 bits (71), Expect = 0.24
Identities = 18/44 (40%), Positives = 24/44 (53%)
Frame = +1
Query: 128 VLILHGQALSSYSYRNLIQSLSTQGVRVIAIDLPGSGFSDKSVE 171
V+ LHG YS+R+ + +L+ G R IA D G G SD E
Sbjct: 112 VVFLHGFPEIWYSWRHQMIALANAGFRAIAPDFRGYGLSDPPPE 243
>AV779571
Length = 538
Score = 31.6 bits (70), Expect = 0.31
Identities = 18/47 (38%), Positives = 26/47 (55%), Gaps = 5/47 (10%)
Frame = +1
Query: 5 TEEQESDQQQQQQKQKQKQK-----QQHSNKTKSKTQKPSTTQPQQT 46
T+ +QQQQQQ+Q+Q Q+ QQ N + +Q S QPQ +
Sbjct: 244 TQNHNHNQQQQQQQQQQTQQQVVDNQQIGNAGSTMSQFVSAPQPQSS 384
>AV415922
Length = 422
Score = 31.6 bits (70), Expect = 0.31
Identities = 14/23 (60%), Positives = 19/23 (81%)
Frame = +3
Query: 7 EQESDQQQQQQKQKQKQKQQHSN 29
+Q QQQQQQ+Q+Q+Q+QQ SN
Sbjct: 174 QQR*QQQQQQQQQQQQQQQQPSN 242
Score = 29.6 bits (65), Expect = 1.2
Identities = 12/27 (44%), Positives = 21/27 (77%)
Frame = +3
Query: 6 EEQESDQQQQQQKQKQKQKQQHSNKTK 32
+ Q+ QQQQQQ+Q+Q+Q+QQ + ++
Sbjct: 168 QHQQR*QQQQQQQQQQQQQQQQPSNSQ 248
>BG662093
Length = 387
Score = 31.2 bits (69), Expect = 0.40
Identities = 17/43 (39%), Positives = 29/43 (66%), Gaps = 1/43 (2%)
Frame = +3
Query: 1 MAIITEEQESDQQQQQQ-KQKQKQKQQHSNKTKSKTQKPSTTQ 42
MAII + E + +++ +K+KQK+ HS+K+K K KPS ++
Sbjct: 123 MAIIRKSVEGKEHKKKDPSKKEKQKKHHSSKSKHK--KPSDSE 245
>TC15910 similar to UP|Q9AR64 (Q9AR64) Urease accessory protein G, partial
(88%)
Length = 1061
Score = 30.8 bits (68), Expect = 0.53
Identities = 11/27 (40%), Positives = 12/27 (43%)
Frame = +2
Query: 458 HDHHQGHSHGHDRLGEAHIHEAGYMDA 484
H HH H H HD +H H G A
Sbjct: 38 HHHHSDHGHDHDHHDNSHSHSDGVASA 118
>TC17520 similar to GB|AAO63388.1|28950929|BT005324 At5g19850 {Arabidopsis
thaliana;}, partial (43%)
Length = 688
Score = 30.4 bits (67), Expect = 0.69
Identities = 17/46 (36%), Positives = 28/46 (59%)
Frame = +1
Query: 123 NPSETVLILHGQALSSYSYRNLIQSLSTQGVRVIAIDLPGSGFSDK 168
N ++++HG +S +R I +L+ + RV +IDL G G+SDK
Sbjct: 277 NDGPALVLVHGFGANSDHWRKNIPALA-KSHRVYSIDLIGYGYSDK 411
>TC7925 homologue to UP|RPE_SPIOL (Q43157) Ribulose-phosphate 3-epimerase,
chloroplast precursor (Pentose-5-phosphate 3-epimerase)
(PPE) (RPE) (R5P3E) , partial (94%)
Length = 1387
Score = 29.6 bits (65), Expect = 1.2
Identities = 17/38 (44%), Positives = 24/38 (62%)
Frame = +3
Query: 13 QQQQQKQKQKQKQQHSNKTKSKTQKPSTTQPQQTNPLS 50
Q+++QKQKQKQKQ+ N+ + + ST Q Q P S
Sbjct: 96 QRKKQKQKQKQKQK-QNEMAATSLCSSTLQSQINGPSS 206
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.317 0.134 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,112,758
Number of Sequences: 28460
Number of extensions: 146020
Number of successful extensions: 1457
Number of sequences better than 10.0: 94
Number of HSP's better than 10.0 without gapping: 1265
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1368
length of query: 494
length of database: 4,897,600
effective HSP length: 94
effective length of query: 400
effective length of database: 2,222,360
effective search space: 888944000
effective search space used: 888944000
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 57 (26.6 bits)
Medicago: description of AC146571.4


