

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146570.9 + phase: 0
(475 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
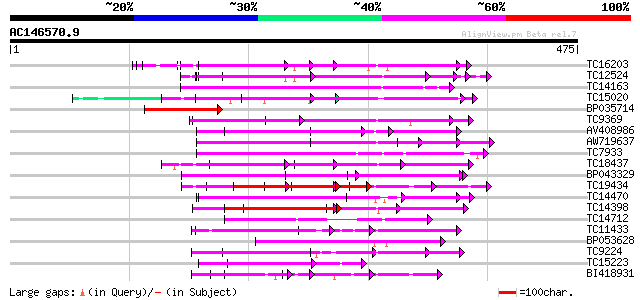
Score E
Sequences producing significant alignments: (bits) Value
TC16203 UP|Q8GRU6 (Q8GRU6) LRR receptor-like kinase (Hypernodula... 119 1e-27
TC12524 similar to UP|Q9LKZ6 (Q9LKZ6) Receptor-like protein kina... 102 1e-22
TC14163 102 2e-22
TC15020 homologue to UP|Q9QX99 (Q9QX99) Transcription factor (T-... 97 4e-21
BP035714 97 6e-21
TC9369 similar to UP|Q93X72 (Q93X72) Leucine-rich repeat resista... 94 5e-20
AV408986 92 1e-19
AW719637 89 2e-18
TC7933 weakly similar to UP|AAQ56728 (AAQ56728) Polygalacturonas... 87 8e-18
TC18437 weakly similar to UP|Q9LVB3 (Q9LVB3) Receptor-like prote... 86 1e-17
BP043329 84 5e-17
TC19434 similar to UP|Q94F63 (Q94F63) Somatic embryogenesis rece... 83 1e-16
TC14470 similar to UP|AAQ56728 (AAQ56728) Polygalacturonase inhi... 82 2e-16
TC14398 similar to UP|Q96477 (Q96477) LRR protein, partial (86%) 78 4e-15
TC14712 similar to UP|PGI2_PHAVU (P58822) Polygalacturonase inhi... 77 6e-15
TC11433 similar to UP|AAR99871 (AAR99871) Strubbelig receptor fa... 73 1e-13
BP053628 71 3e-13
TC9224 similar to GB|AAP21230.1|30102624|BT006422 At3g49750 {Ara... 70 6e-13
TC15223 similar to UP|CAE76632 (CAE76632) Leucine rich repeat pr... 69 2e-12
BI418931 69 2e-12
>TC16203 UP|Q8GRU6 (Q8GRU6) LRR receptor-like kinase (Hypernodulation
aberrant root formation protein), complete
Length = 3308
Score = 119 bits (297), Expect = 1e-27
Identities = 78/231 (33%), Positives = 124/231 (52%), Gaps = 2/231 (0%)
Frame = +2
Query: 159 GNIPSTFGV-LKNLQSLVLLENGLTGNIPQEIGNLVKLKRLVLSGNNFSGNIPDIFGGLS 217
G P V + L++L +N +G +P+EI L KLK L L+GN FSG IP+ +
Sbjct: 515 GQFPGNITVGMTELEALDAYDNSFSGPLPEEIVKLEKLKYLHLAGNYFSGTIPESYSEFQ 694
Query: 218 DLLILDLSRNSLSGTLPVTLGRLISVLKLDLSH-NFLEGKLLNEFGNLKNLTLMDLRNNR 276
L L L+ NSL+G +P +L +L ++ +L L + N EG + FG+++NL L+++ N
Sbjct: 695 SLEFLGLNANSLTGRVPESLAKLKTLKELHLGYSNAYEGGIPPAFGSMENLRLLEMANCN 874
Query: 277 LCCGLVLSLQEMNSLEEMVLSNNPLGGDIRTLKWENLQNLVILELSNMELIGEIPESLSQ 336
L + SL + L + + N L G I + ++ +L+ L+LS +L GEIPES S+
Sbjct: 875 LTGEIPPSLGNLTKLHSLFVQMNNLTGTIPP-ELSSMMSLMSLDLSINDLTGEIPESFSK 1051
Query: 337 LKKLRFLGLSDNNITGNLSPKLETLPSLNALYLSGNNLKGEIQFSKGFFGK 387
LK L + N G+L + LP+L L + NN + + G G+
Sbjct: 1052LKNLTLMNFFQNKFRGSLPSFIGDLPNLETLQVWENNFSFVLPHNLGGNGR 1204
Score = 115 bits (288), Expect = 2e-26
Identities = 96/299 (32%), Positives = 142/299 (47%), Gaps = 24/299 (8%)
Frame = +2
Query: 104 EFKPELFQLKHLKAISFFNCFQSPNKLPVSIPTGNWEKLAESLESIEFRSNPGLIGNIPS 163
E P L L L ++ F N L +IP + SL S++ N L G IP
Sbjct: 884 EIPPSLGNLTKLHSL-----FVQMNNLTGTIPPELSSMM--SLMSLDLSIND-LTGEIPE 1039
Query: 164 TFGVLKNLQSLVLLENGLTGNIPQEIGNLVKLKRLVLSGNNFSGNIPDIFGGLSDLLILD 223
+F LKNL + +N G++P IG+L L+ L + NNFS +P GG L D
Sbjct: 1040 SFSKLKNLTLMNFFQNKFRGSLPSFIGDLPNLETLQVWENNFSFVLPHNLGGNGRFLYFD 1219
Query: 224 LSRNSLSGTLPVTL------------------------GRLISVLKLDLSHNFLEGKLLN 259
+++N L+G +P L G S+ K+ +++NFL+G +
Sbjct: 1220 VTKNHLTGLIPPDLCKSGRLKTFIITDNFFRGPIPKGIGECRSLTKIRVANNFLDGPVPP 1399
Query: 260 EFGNLKNLTLMDLRNNRLCCGLVLSLQEMNSLEEMVLSNNPLGGDIRTLKWENLQNLVIL 319
L ++T+ +L NNRL G + S+ SL + LSNN G I +NL+ L L
Sbjct: 1400 GVFQLPSVTITELSNNRL-NGELPSVISGESLGTLTLSNNLFTGKI-PAAMKNLRALQSL 1573
Query: 320 ELSNMELIGEIPESLSQLKKLRFLGLSDNNITGNLSPKLETLPSLNALYLSGNNLKGEI 378
L E IGEIP + ++ L + +S NN+TG + + SL A+ LS NNL GE+
Sbjct: 1574 SLDANEFIGEIPGGVFEIPMLTKVNISGNNLTGPIPTTITHRASLTAVDLSRNNLAGEV 1750
Score = 107 bits (266), Expect = 6e-24
Identities = 79/259 (30%), Positives = 129/259 (49%), Gaps = 24/259 (9%)
Frame = +2
Query: 144 ESLESIEFRSNPGLIGNIPSTFGVLKNLQSLVL-LENGLTGNIPQEIGNLVKLKRLVLSG 202
+SLE + +N L G +P + LK L+ L L N G IP G++ L+ L ++
Sbjct: 692 QSLEFLGLNAN-SLTGRVPESLAKLKTLKELHLGYSNAYEGGIPPAFGSMENLRLLEMAN 868
Query: 203 NNFSGNIPDIFGGLSDLLILDLSRNSLSGTLPVTLGRLISVLKLDLSHNFLEGKLLNEFG 262
N +G IP G L+ L L + N+L+GT+P L ++S++ LDLS N L G++ F
Sbjct: 869 CNLTGEIPPSLGNLTKLHSLFVQMNNLTGTIPPELSSMMSLMSLDLSINDLTGEIPESFS 1048
Query: 263 NLKNLTLMDLRNNRLCCGLVLSLQEMNSLEEM-VLSNN-------PLGGDIRTLKWENLQ 314
LKNLTLM+ N+ L + ++ +LE + V NN LGG+ R L ++ +
Sbjct: 1049KLKNLTLMNFFQNKFRGSLPSFIGDLPNLETLQVWENNFSFVLPHNLGGNGRFLYFDVTK 1228
Query: 315 N---------------LVILELSNMELIGEIPESLSQLKKLRFLGLSDNNITGNLSPKLE 359
N L +++ G IP+ + + + L + +++N + G + P +
Sbjct: 1229NHLTGLIPPDLCKSGRLKTFIITDNFFRGPIPKGIGECRSLTKIRVANNFLDGPVPPGVF 1408
Query: 360 TLPSLNALYLSGNNLKGEI 378
LPS+ LS N L GE+
Sbjct: 1409QLPSVTITELSNNRLNGEL 1465
Score = 74.3 bits (181), Expect = 4e-14
Identities = 55/169 (32%), Positives = 86/169 (50%)
Frame = +2
Query: 107 PELFQLKHLKAISFFNCFQSPNKLPVSIPTGNWEKLAESLESIEFRSNPGLIGNIPSTFG 166
P +FQL + N N+L +P+ ESL ++ SN G IP+
Sbjct: 1397 PGVFQLPSVTITELSN-----NRLNGELPS---VISGESLGTLTL-SNNLFTGKIPAAMK 1549
Query: 167 VLKNLQSLVLLENGLTGNIPQEIGNLVKLKRLVLSGNNFSGNIPDIFGGLSDLLILDLSR 226
L+ LQSL L N G IP + + L ++ +SGNN +G IP + L +DLSR
Sbjct: 1550 NLRALQSLSLDANEFIGEIPGGVFEIPMLTKVNISGNNLTGPIPTTITHRASLTAVDLSR 1729
Query: 227 NSLSGTLPVTLGRLISVLKLDLSHNFLEGKLLNEFGNLKNLTLMDLRNN 275
N+L+G +P + L+ + L+LS N + G + +E + +LT +DL +N
Sbjct: 1730 NNLAGEVPKGMKNLMDLSILNLSRNEISGPVPDEIRFMTSLTTLDLSSN 1876
Score = 63.9 bits (154), Expect = 5e-11
Identities = 42/115 (36%), Positives = 59/115 (50%)
Frame = +2
Query: 141 KLAESLESIEFRSNPGLIGNIPSTFGVLKNLQSLVLLENGLTGNIPQEIGNLVKLKRLVL 200
K +L+S+ +N IG IP + L + + N LTG IP I + L + L
Sbjct: 1547 KNLRALQSLSLDANE-FIGEIPGGVFEIPMLTKVNISGNNLTGPIPTTITHRASLTAVDL 1723
Query: 201 SGNNFSGNIPDIFGGLSDLLILDLSRNSLSGTLPVTLGRLISVLKLDLSHNFLEG 255
S NN +G +P L DL IL+LSRN +SG +P + + S+ LDLS N G
Sbjct: 1724 SRNNLAGEVPKGMKNLMDLSILNLSRNEISGPVPDEIRFMTSLTTLDLSSNNFTG 1888
Score = 58.5 bits (140), Expect = 2e-09
Identities = 41/123 (33%), Positives = 63/123 (50%)
Frame = +2
Query: 112 LKHLKAISFFNCFQSPNKLPVSIPTGNWEKLAESLESIEFRSNPGLIGNIPSTFGVLKNL 171
+K+L+A+ + N+ IP G +E L + N L G IP+T +L
Sbjct: 1544 MKNLRALQSLSL--DANEFIGEIPGGVFE--IPMLTKVNISGN-NLTGPIPTTITHRASL 1708
Query: 172 QSLVLLENGLTGNIPQEIGNLVKLKRLVLSGNNFSGNIPDIFGGLSDLLILDLSRNSLSG 231
++ L N L G +P+ + NL+ L L LS N SG +PD ++ L LDLS N+ +G
Sbjct: 1709 TAVDLSRNNLAGEVPKGMKNLMDLSILNLSRNEISGPVPDEIRFMTSLTTLDLSSNNFTG 1888
Query: 232 TLP 234
T+P
Sbjct: 1889 TVP 1897
Score = 37.7 bits (86), Expect = 0.004
Identities = 28/102 (27%), Positives = 50/102 (48%), Gaps = 1/102 (0%)
Frame = +2
Query: 288 MNSLEEMVLSNNPLGGDIRTLKWENLQNLVILELSNMELIGEIPESLSQ-LKKLRFLGLS 346
+ LE + +S N L + + +L +L +L +S+ G+ P +++ + +L L
Sbjct: 398 LEKLENLTISMNNLTDQLPS-DLASLTSLKVLNISHNLFSGQFPGNITVGMTELEALDAY 574
Query: 347 DNNITGNLSPKLETLPSLNALYLSGNNLKGEIQFSKGFFGKL 388
DN+ +G L ++ L L L+L+GN G I S F L
Sbjct: 575 DNSFSGPLPEEIVKLEKLKYLHLAGNYFSGTIPESYSEFQSL 700
>TC12524 similar to UP|Q9LKZ6 (Q9LKZ6) Receptor-like protein kinase 1,
partial (27%)
Length = 834
Score = 102 bits (254), Expect = 1e-22
Identities = 76/208 (36%), Positives = 104/208 (49%)
Frame = +2
Query: 179 NGLTGNIPQEIGNLVKLKRLVLSGNNFSGNIPDIFGGLSDLLILDLSRNSLSGTLPVTLG 238
N G IP EIGNL +L R + SG IP G L L L L N LSG+L LG
Sbjct: 23 NNYQGGIPPEIGNLTQLLRFDAAYCGLSGEIPAELGKLQKLDTLFLQVNVLSGSLTPELG 202
Query: 239 RLISVLKLDLSHNFLEGKLLNEFGNLKNLTLMDLRNNRLCCGLVLSLQEMNSLEEMVLSN 298
L S+ +DLS+N L G++ F LKNLTL++L NRL + + EM +LE + L
Sbjct: 203 HLKSLKSMDLSNNMLSGQVPASFAELKNLTLLNLFRNRLHGAIPEFVGEMPALEVLQLWE 382
Query: 299 NPLGGDIRTLKWENLQNLVILELSNMELIGEIPESLSQLKKLRFLGLSDNNITGNLSPKL 358
N G I +N L +++LS+ +L G +P + +L+ L N + G + L
Sbjct: 383 NNFTGSIPQSLGKN-GKLTLVDLSSNKLTGTLPPHMCSGNRLQTLIALGNFLFGPIPESL 559
Query: 359 ETLPSLNALYLSGNNLKGEIQFSKGFFG 386
SL + + N L G I KG FG
Sbjct: 560 GKCESLTRIRMGQNFLNGSI--PKGLFG 637
Score = 97.1 bits (240), Expect = 6e-21
Identities = 76/245 (31%), Positives = 109/245 (44%)
Frame = +2
Query: 159 GNIPSTFGVLKNLQSLVLLENGLTGNIPQEIGNLVKLKRLVLSGNNFSGNIPDIFGGLSD 218
G IP G L L GL+G IP E+G L KL L L N SG++ G L
Sbjct: 35 GGIPPEIGNLTQLLRFDAAYCGLSGEIPAELGKLQKLDTLFLQVNVLSGSLTPELGHLKS 214
Query: 219 LLILDLSRNSLSGTLPVTLGRLISVLKLDLSHNFLEGKLLNEFGNLKNLTLMDLRNNRLC 278
L +DLS N LSG +P + L ++ L+L N L G + G + L ++ L N
Sbjct: 215 LKSMDLSNNMLSGQVPASFAELKNLTLLNLFRNRLHGAIPEFVGEMPALEVLQLWENNFT 394
Query: 279 CGLVLSLQEMNSLEEMVLSNNPLGGDIRTLKWENLQNLVILELSNMELIGEIPESLSQLK 338
+ SL + L + LS+N L G + + ++ L N L G IPESL + +
Sbjct: 395 GSIPQSLGKNGKLTLVDLSSNKLTGTLPPHMCSGNRLQTLIALGNF-LFGPIPESLGKCE 571
Query: 339 KLRFLGLSDNNITGNLSPKLETLPSLNALYLSGNNLKGEIQFSKGFFGKLGRRFGAWSNP 398
L + + N + G++ L LP L + N L GE + +G+ SN
Sbjct: 572 SLTRIRMGQNFLNGSIPKGLFGLPKLTQVEFQDNLLSGEFPETGSVSHNIGQI--TLSNN 745
Query: 399 KLCYP 403
KL P
Sbjct: 746 KLSGP 760
Score = 93.6 bits (231), Expect = 6e-20
Identities = 69/212 (32%), Positives = 110/212 (51%), Gaps = 3/212 (1%)
Frame = +2
Query: 144 ESLESIEFRSNPGLIGNIPSTFGVLKNLQSLVLLENGLTGNIPQEIGNLVKLKRLVLSGN 203
+SL+S++ SN L G +P++F LKNL L L N L G IP+ +G + L+ L L N
Sbjct: 209 KSLKSMDL-SNNMLSGQVPASFAELKNLTLLNLFRNRLHGAIPEFVGEMPALEVLQLWEN 385
Query: 204 NFSGNIPDIFGGLSDLLILDLSRNSLSGTLPVTL---GRLISVLKLDLSHNFLEGKLLNE 260
NF+G+IP G L ++DLS N L+GTLP + RL +++ L NFL G +
Sbjct: 386 NFTGSIPQSLGKNGKLTLVDLSSNKLTGTLPPHMCSGNRLQTLIALG---NFLFGPIPES 556
Query: 261 FGNLKNLTLMDLRNNRLCCGLVLSLQEMNSLEEMVLSNNPLGGDIRTLKWENLQNLVILE 320
G ++LT + + N L + L + L ++ +N L G+ + N+ +
Sbjct: 557 LGKCESLTRIRMGQNFLNGSIPKGLFGLPKLTQVEFQDNLLSGEFPETGSVS-HNIGQIT 733
Query: 321 LSNMELIGEIPESLSQLKKLRFLGLSDNNITG 352
LSN +L G +P ++ ++ L L N +G
Sbjct: 734 LSNNKLSGPLPSTIGNFTSMQKLLLDGNKFSG 829
Score = 91.3 bits (225), Expect = 3e-19
Identities = 74/245 (30%), Positives = 111/245 (45%), Gaps = 24/245 (9%)
Frame = +2
Query: 156 GLIGNIPSTFGVLKNLQSLVLLENGLTGNIPQEIGNLVKLKRLVLSGNNFSGNIPDIFGG 215
GL G IP+ G L+ L +L L N L+G++ E+G+L LK + LS N SG +P F
Sbjct: 98 GLSGEIPAELGKLQKLDTLFLQVNVLSGSLTPELGHLKSLKSMDLSNNMLSGQVPASFAE 277
Query: 216 LSDLLILDLSRNSL------------------------SGTLPVTLGRLISVLKLDLSHN 251
L +L +L+L RN L +G++P +LG+ + +DLS N
Sbjct: 278 LKNLTLLNLFRNRLHGAIPEFVGEMPALEVLQLWENNFTGSIPQSLGKNGKLTLVDLSSN 457
Query: 252 FLEGKLLNEFGNLKNLTLMDLRNNRLCCGLVLSLQEMNSLEEMVLSNNPLGGDIRTLKWE 311
L G L + L + N L + SL + SL + + N L G I +
Sbjct: 458 KLTGTLPPHMCSGNRLQTLIALGNFLFGPIPESLGKCESLTRIRMGQNFLNGSIPKGLF- 634
Query: 312 NLQNLVILELSNMELIGEIPESLSQLKKLRFLGLSDNNITGNLSPKLETLPSLNALYLSG 371
L L +E + L GE PE+ S + + LS+N ++G L + S+ L L G
Sbjct: 635 GLPKLTQVEFQDNLLSGEFPETGSVSHNIGQITLSNNKLSGPLPSTIGNFTSMQKLLLDG 814
Query: 372 NNLKG 376
N G
Sbjct: 815 NKFSG 829
Score = 57.0 bits (136), Expect = 7e-09
Identities = 34/100 (34%), Positives = 52/100 (52%)
Frame = +2
Query: 157 LIGNIPSTFGVLKNLQSLVLLENGLTGNIPQEIGNLVKLKRLVLSGNNFSGNIPDIFGGL 216
L G IP + G ++L + + +N L G+IP+ + L KL ++ N SG P+
Sbjct: 533 LFGPIPESLGKCESLTRIRMGQNFLNGSIPKGLFGLPKLTQVEFQDNLLSGEFPETGSVS 712
Query: 217 SDLLILDLSRNSLSGTLPVTLGRLISVLKLDLSHNFLEGK 256
++ + LS N LSG LP T+G S+ KL L N G+
Sbjct: 713 HNIGQITLSNNKLSGPLPSTIGNFTSMQKLLLDGNKFSGR 832
>TC14163
Length = 1712
Score = 102 bits (253), Expect = 2e-22
Identities = 72/230 (31%), Positives = 116/230 (50%), Gaps = 1/230 (0%)
Frame = +1
Query: 144 ESLESIEFRSNPGLIGNIPSTFGVLKNLQSLVLLENGLTGNIPQEIGNLVKLKRLVLSGN 203
++L+ + + G P L LQ + + N L+G IP+ IGNL +L L L+GN
Sbjct: 412 KNLDGFYLLNLKNISGPFPGFLFKLPKLQFIYIENNQLSGRIPENIGNLTRLDVLSLTGN 591
Query: 204 NFSGNIPDIFGGLSDLLILDLSRNSLSGTLPVTLGRLISVLKLDLSHNFLEGKLLNEFGN 263
F+G IP GGL+ L L L NSL+GT+P T+ RL ++ L L N G + + F +
Sbjct: 592 RFTGTIPSSVGGLTHLTQLQLGNNSLTGTIPATIARLKNLTYLSLEGNQFSGAIPDFFSS 771
Query: 264 LKNLTLMDLRNNRLCCGLVLSLQEM-NSLEEMVLSNNPLGGDIRTLKWENLQNLVILELS 322
+L ++ L N+ + S+ + L + L +N L G I + + L L+LS
Sbjct: 772 FTDLGILRLSRNKFSGKIPASISTLAPKLRYLELGHNQLSGKIPDFLGK-FRALDTLDLS 948
Query: 323 NMELIGEIPESLSQLKKLRFLGLSDNNITGNLSPKLETLPSLNALYLSGN 372
+ G +P S L K+ L L+ NN+ + P++ + + +L LS N
Sbjct: 949 SNRFSGTVPASFKNLTKIFNLNLA-NNLLVDPFPEM-NVKGIESLDLSNN 1092
>TC15020 homologue to UP|Q9QX99 (Q9QX99) Transcription factor (T-cell
leukemia, homeobox 1), partial (5%)
Length = 1304
Score = 97.4 bits (241), Expect = 4e-21
Identities = 79/240 (32%), Positives = 116/240 (47%), Gaps = 3/240 (1%)
Frame = +1
Query: 156 GLIGNIPSTFGVLKNLQSLVLLENGLTGNIPQEIGNLVKLKRLVLSGNNFSGNIPDIFGG 215
G G++ L L +L L EN +G IP + +L LK L L N+FSG+IP
Sbjct: 262 GYSGSLTPLISQLTQLVTLDLAENSFSGPIPSSLSSLSNLKTLTLRSNSFSGSIPPSITT 441
Query: 216 LSDLLILDLSRNSLSGTLPVTLGRLISVLKLDLSHNFLEGKLLNEFGNLKNLTLMDLRNN 275
L L LD S NSLSG+LP ++ LI++ ++DLS N L G + NL L ++ N
Sbjct: 442 LKSLYSLDFSHNSLSGSLPNSMNSLINLRRIDLSFNKLTGSIPKLPPNLLELA---VKAN 612
Query: 276 RLCCGL-VLSLQEMNSLEEMVLSNNPLGGDIRTLKWENLQNLVILELSNMELIG-EIPE- 332
L L + + +N LE + LS N L G + + L +L ++LSN G +I
Sbjct: 613 SLSGPLQKATFEGLNQLEVVELSENTLAGTVEPW-FFLLPSLQQVDLSNNSFTGVQISRP 789
Query: 333 SLSQLKKLRFLGLSDNNITGNLSPKLETLPSLNALYLSGNNLKGEIQFSKGFFGKLGRRF 392
+ + L + L N I G L P L++L + N+L+G I G + R F
Sbjct: 790 ARGEGSNLVAVNLGFNRIQGYAPANLAAYPQLSSLSIRHNSLRGAIPLEYGGIKSMKRLF 969
Score = 79.7 bits (195), Expect = 9e-16
Identities = 57/189 (30%), Positives = 94/189 (49%), Gaps = 1/189 (0%)
Frame = +1
Query: 195 LKRLVLSGNNFSGNIPDIFGGLSDLLILDLSRNSLSGTLPVTLGRLISVLKLDLSHNFLE 254
+ ++ L +SG++ + L+ L+ LDL+ NS SG +P +L L ++ L L N
Sbjct: 235 INQITLDPAGYSGSLTPLISQLTQLVTLDLAENSFSGPIPSSLSSLSNLKTLTLRSNSFS 414
Query: 255 GKLLNEFGNLKNLTLMDLRNNRLCCGLVLSLQEMNSLEEMVLSNNPLGGDIRTLKWENLQ 314
G + LK+L +D +N L L S+ + +L + LS N L G I L
Sbjct: 415 GSIPPSITTLKSLYSLDFSHNSLSGSLPNSMNSLINLRRIDLSFNKLTGSIPKLP----P 582
Query: 315 NLVILELSNMELIGEIPESLSQ-LKKLRFLGLSDNNITGNLSPKLETLPSLNALYLSGNN 373
NL+ L + L G + ++ + L +L + LS+N + G + P LPSL + LS N+
Sbjct: 583 NLLELAVKANSLSGPLQKATFEGLNQLEVVELSENTLAGTVEPWFFLLPSLQQVDLSNNS 762
Query: 374 LKGEIQFSK 382
G +Q S+
Sbjct: 763 FTG-VQISR 786
Score = 60.5 bits (145), Expect = 6e-10
Identities = 71/298 (23%), Positives = 114/298 (37%), Gaps = 73/298 (24%)
Frame = +1
Query: 53 SWNGSDLYPDPCGSTSIEGVSCDIFNGLWYVTVINIGPIHENSLPCANEKLEFKPELFQL 112
SWN + DPC C GL I++ +L A P + QL
Sbjct: 142 SWNFTI---DPCSLPRRTHFIC----GLTCTATTTTNNINQITLDPAGYSGSLTPLISQL 300
Query: 113 KHLKAISFF-NCFQSPNKLPVSIPTGNWEKLAESLESIEFRSNPGLIGNIPSTFGVLKNL 171
L + N F P +P S+ + +L+++ RSN G+IP + LK+L
Sbjct: 301 TQLVTLDLAENSFSGP--IPSSLSS------LSNLKTLTLRSN-SFSGSIPPSITTLKSL 453
Query: 172 QSLVLLENGLTGNIPQEIGNLVKLKRLVLSGNNFSGNIPDI------------------- 212
SL N L+G++P + +L+ L+R+ LS N +G+IP +
Sbjct: 454 YSLDFSHNSLSGSLPNSMNSLINLRRIDLSFNKLTGSIPKLPPNLLELAVKANSLSGPLQ 633
Query: 213 ---FGGLSDLLILDLSRNSLSGTLPVTLGRLISVLKLDLS-------------------- 249
F GL+ L +++LS N+L+GT+ L S+ ++DLS
Sbjct: 634 KATFEGLNQLEVVELSENTLAGTVEPWFFLLPSLQQVDLSNNSFTGVQISRPARGEGSNL 813
Query: 250 ------------------------------HNFLEGKLLNEFGNLKNLTLMDLRNNRL 277
HN L G + E+G +K++ + L N L
Sbjct: 814 VAVNLGFNRIQGYAPANLAAYPQLSSLSIRHNSLRGAIPLEYGGIKSMKRLFLDGNFL 987
Score = 47.0 bits (110), Expect = 7e-06
Identities = 42/134 (31%), Positives = 58/134 (42%), Gaps = 5/134 (3%)
Frame = +1
Query: 128 NKLPVSIPTGNWEKLAESLESIEFRSNPGLIGNIPSTFGVLKNLQSLVLLENGLTG---- 183
N L + +E L + LE +E N L G + F +L +LQ + L N TG
Sbjct: 610 NSLSGPLQKATFEGLNQ-LEVVELSENT-LAGTVEPWFFLLPSLQQVDLSNNSFTGVQIS 783
Query: 184 -NIPQEIGNLVKLKRLVLSGNNFSGNIPDIFGGLSDLLILDLSRNSLSGTLPVTLGRLIS 242
E NLV + L N G P L L + NSL G +P+ G + S
Sbjct: 784 RPARGEGSNLVAVN---LGFNRIQGYAPANLAAYPQLSSLSIRHNSLRGAIPLEYGGIKS 954
Query: 243 VLKLDLSHNFLEGK 256
+ +L L NFL+GK
Sbjct: 955 MKRLFLDGNFLDGK 996
>BP035714
Length = 391
Score = 97.1 bits (240), Expect = 6e-21
Identities = 44/65 (67%), Positives = 56/65 (85%)
Frame = -2
Query: 114 HLKAISFFNCFQSPNKLPVSIPTGNWEKLAESLESIEFRSNPGLIGNIPSTFGVLKNLQS 173
HLKA+SF CF+S ++ PV+IP GNWEKLA +LES+EFRSN GL G IPS+FGVL+NLQS
Sbjct: 390 HLKALSFIKCFESQHRNPVTIPNGNWEKLAGTLESLEFRSNNGLTGKIPSSFGVLRNLQS 211
Query: 174 LVLLE 178
L+L++
Sbjct: 210 LLLID 196
>TC9369 similar to UP|Q93X72 (Q93X72) Leucine-rich repeat resistance
protein-like protein, partial (70%)
Length = 859
Score = 94.0 bits (232), Expect = 5e-20
Identities = 75/219 (34%), Positives = 98/219 (44%)
Frame = +1
Query: 154 NPGLIGNIPSTFGVLKNLQSLVLLENGLTGNIPQEIGNLVKLKRLVLSGNNFSGNIPDIF 213
N L G IP G LK L+ L L N L IP EIG L L L LS N+F G IP
Sbjct: 34 NNKLTGPIPPQIGRLKRLKILNLRWNKLQDAIPPEIGELKSLTHLYLSFNSFKGEIPKEL 213
Query: 214 GGLSDLLILDLSRNSLSGTLPVTLGRLISVLKLDLSHNFLEGKLLNEFGNLKNLTLMDLR 273
L DL L L N L G +P LG L ++ LD +N L G T+ +L
Sbjct: 214 ANLPDLRYLYLHENRLIGRIPPELGTLQNLRHLDAGNNHLVG------------TIRELI 357
Query: 274 NNRLCCGLVLSLQEMNSLEEMVLSNNPLGGDIRTLKWENLQNLVILELSNMELIGEIPES 333
C +L + L+NN G + + NL +L IL LS ++ G IP S
Sbjct: 358 RIEGC---------FPALRNLYLNNNYFTGGM-PAQLANLSSLEILYLSYNKMSGVIPSS 507
Query: 334 LSQLKKLRFLGLSDNNITGNLSPKLETLPSLNALYLSGN 372
L+ + KL +L L N +G + L +Y+ GN
Sbjct: 508 LAHIPKLTYLYLDHNQFSGRIPEPFYKHSFLKEMYIEGN 624
Score = 77.4 bits (189), Expect = 5e-15
Identities = 63/177 (35%), Positives = 84/177 (46%), Gaps = 3/177 (1%)
Frame = +1
Query: 215 GLSDLLILDLSRNSLSGTLPVTLGRLISVLKLDLSHNFLEGKLLNEFGNLKNLTLMDLRN 274
G DL LDL N L+G +P +GRL + L+L N L+ + E G LK+LT + L
Sbjct: 1 GTRDLTRLDLHNNKLTGPIPPQIGRLKRLKILNLRWNKLQDAIPPEIGELKSLTHLYLSF 180
Query: 275 NRLCCGLVLSLQEMNSLEEMVLSNNPLGGDIRTLKWENLQNLVILELSNMELIGEIPESL 334
N + L + L + L N L G I + LQNL L+ N L+G I E +
Sbjct: 181 NSFKGEIPKELANLPDLRYLYLHENRLIGRIPP-ELGTLQNLRHLDAGNNHLVGTIRELI 357
Query: 335 ---SQLKKLRFLGLSDNNITGNLSPKLETLPSLNALYLSGNNLKGEIQFSKGFFGKL 388
LR L L++N TG + +L L SL LYLS N + G I S KL
Sbjct: 358 RIEGCFPALRNLYLNNNYFTGGMPAQLANLSSLEILYLSYNKMSGVIPSSLAHIPKL 528
Score = 44.3 bits (103), Expect = 4e-05
Identities = 29/98 (29%), Positives = 48/98 (48%), Gaps = 1/98 (1%)
Frame = +1
Query: 151 FRSNPGLIGNIPSTFGVLKNLQSLVLLENGLTGNIPQEIGNLVKLKRLVLSGNNFSGNIP 210
+ +N G +P+ L +L+ L L N ++G IP + ++ KL L L N FSG IP
Sbjct: 394 YLNNNYFTGGMPAQLANLSSLEILYLSYNKMSGVIPSSLAHIPKLTYLYLDHNQFSGRIP 573
Query: 211 DIFGGLSDLLILDLSRNSL-SGTLPVTLGRLISVLKLD 247
+ F S L + + N+ G P+ +++ V D
Sbjct: 574 EPFYKHSFLKEMYIEGNAFRPGVKPIGFHKVLEVSDSD 687
>AV408986
Length = 434
Score = 92.4 bits (228), Expect = 1e-19
Identities = 51/143 (35%), Positives = 80/143 (55%)
Frame = +3
Query: 157 LIGNIPSTFGVLKNLQSLVLLENGLTGNIPQEIGNLVKLKRLVLSGNNFSGNIPDIFGGL 216
L G IP G + +L+ L L +N +G IP+E+G L LKRL + N +G IP G
Sbjct: 3 LSGEIPPEIGNISSLELLALHQNSFSGAIPKELGKLSGLKRLYVYTNQLNGTIPTELGNC 182
Query: 217 SDLLILDLSRNSLSGTLPVTLGRLISVLKLDLSHNFLEGKLLNEFGNLKNLTLMDLRNNR 276
++ + +DLS N L G +P LG++ ++ L L N L+G + E G+L+ L +DL N
Sbjct: 183 TNAIEIDLSENRLIGIIPKELGQISNLSLLHLFENNLQGHIPRELGSLRQLKKLDLSLNN 362
Query: 277 LCCGLVLSLQEMNSLEEMVLSNN 299
L + L Q + +E++ L +N
Sbjct: 363 LTGTIPLEFQNLTYIEDLQLFDN 431
Score = 89.0 bits (219), Expect = 2e-18
Identities = 56/141 (39%), Positives = 81/141 (56%)
Frame = +3
Query: 181 LTGNIPQEIGNLVKLKRLVLSGNNFSGNIPDIFGGLSDLLILDLSRNSLSGTLPVTLGRL 240
L+G IP EIGN+ L+ L L N+FSG IP G LS L L + N L+GT+P LG
Sbjct: 3 LSGEIPPEIGNISSLELLALHQNSFSGAIPKELGKLSGLKRLYVYTNQLNGTIPTELGNC 182
Query: 241 ISVLKLDLSHNFLEGKLLNEFGNLKNLTLMDLRNNRLCCGLVLSLQEMNSLEEMVLSNNP 300
+ +++DLS N L G + E G + NL+L+ L N L + L + L+++ LS N
Sbjct: 183 TNAIEIDLSENRLIGIIPKELGQISNLSLLHLFENNLQGHIPRELGSLRQLKKLDLSLNN 362
Query: 301 LGGDIRTLKWENLQNLVILEL 321
L G I L+++NL + L+L
Sbjct: 363 LTGTI-PLEFQNLTYIEDLQL 422
Score = 71.6 bits (174), Expect = 3e-13
Identities = 44/126 (34%), Positives = 68/126 (53%)
Frame = +3
Query: 253 LEGKLLNEFGNLKNLTLMDLRNNRLCCGLVLSLQEMNSLEEMVLSNNPLGGDIRTLKWEN 312
L G++ E GN+ +L L+ L N + L +++ L+ + + N L G I T + N
Sbjct: 3 LSGEIPPEIGNISSLELLALHQNSFSGAIPKELGKLSGLKRLYVYTNQLNGTIPT-ELGN 179
Query: 313 LQNLVILELSNMELIGEIPESLSQLKKLRFLGLSDNNITGNLSPKLETLPSLNALYLSGN 372
N + ++LS LIG IP+ L Q+ L L L +NN+ G++ +L +L L L LS N
Sbjct: 180 CTNAIEIDLSENRLIGIIPKELGQISNLSLLHLFENNLQGHIPRELGSLRQLKKLDLSLN 359
Query: 373 NLKGEI 378
NL G I
Sbjct: 360 NLTGTI 377
>AW719637
Length = 571
Score = 88.6 bits (218), Expect = 2e-18
Identities = 61/191 (31%), Positives = 90/191 (46%), Gaps = 1/191 (0%)
Frame = +2
Query: 157 LIGNIPSTFGVLKNLQSLVLLENGLTGNIPQEIGNLVKLKRLVLSGNNFSGNIPDIFGGL 216
L+G IP L NLQSL L N L GNIP IGNL L L L N+ SG IP G L
Sbjct: 2 LLGEIPEEICSLSNLQSLSLHTNFLEGNIPSNIGNLSSLVNLTLFDNHLSGEIPKSIGSL 181
Query: 217 SDLLILDLSRN-SLSGTLPVTLGRLISVLKLDLSHNFLEGKLLNEFGNLKNLTLMDLRNN 275
+L + N +L G +P +G S++ L L+ + G L + LK + + +
Sbjct: 182 RNLQVFRAGGNKNLKGEIPWEIGNCTSLVMLGLAETSISGSLPSSIQLLKRIKTIAIYTT 361
Query: 276 RLCCGLVLSLQEMNSLEEMVLSNNPLGGDIRTLKWENLQNLVILELSNMELIGEIPESLS 335
L + + + L+ + L N + G I + E L L L L ++G IPE +
Sbjct: 362 LLSGSIPEEIGNCSELQNLYLYQNSISGSIPSQIGE-LSKLKSLLLWQNNIVGTIPEEIG 538
Query: 336 QLKKLRFLGLS 346
+ ++ + LS
Sbjct: 539 RCTEMEVIDLS 571
Score = 50.1 bits (118), Expect = 8e-07
Identities = 36/127 (28%), Positives = 62/127 (48%), Gaps = 1/127 (0%)
Frame = +2
Query: 253 LEGKLLNEFGNLKNLTLMDLRNNRLCCGLVLSLQEMNSLEEMVLSNNPLGGDIRTLKWEN 312
L G++ E +L NL + L N L + ++ ++SL + L +N L G+I +
Sbjct: 2 LLGEIPEEICSLSNLQSLSLHTNFLEGNIPSNIGNLSSLVNLTLFDNHLSGEIPK-SIGS 178
Query: 313 LQNLVILELS-NMELIGEIPESLSQLKKLRFLGLSDNNITGNLSPKLETLPSLNALYLSG 371
L+NL + N L GEIP + L LGL++ +I+G+L ++ L + + +
Sbjct: 179 LRNLQVFRAGGNKNLKGEIPWEIGNCTSLVMLGLAETSISGSLPSSIQLLKRIKTIAIYT 358
Query: 372 NNLKGEI 378
L G I
Sbjct: 359 TLLSGSI 379
Score = 44.3 bits (103), Expect = 4e-05
Identities = 32/82 (39%), Positives = 41/82 (49%), Gaps = 1/82 (1%)
Frame = +2
Query: 326 LIGEIPESLSQLKKLRFLGLSDNNITGNLSPKLETLPSLNALYLSGNNLKGEIQFSKGFF 385
L+GEIPE + L L+ L L N + GN+ + L SL L L N+L GEI S G
Sbjct: 2 LLGEIPEEICSLSNLQSLSLHTNFLEGNIPSNIGNLSSLVNLTLFDNHLSGEIPKSIGSL 181
Query: 386 GKLG-RRFGAWSNPKLCYPFEL 406
L R G N K P+E+
Sbjct: 182 RNLQVFRAGGNKNLKGEIPWEI 247
>TC7933 weakly similar to UP|AAQ56728 (AAQ56728) Polygalacturonase
inhibiting protein, partial (78%)
Length = 1320
Score = 86.7 bits (213), Expect = 8e-18
Identities = 83/249 (33%), Positives = 123/249 (49%), Gaps = 4/249 (1%)
Frame = +3
Query: 157 LIGNIPSTFGVLKNLQSLVLLE-NGLTGNIPQEIGNLVKLKRLVLSGNNFSGNIPDIFGG 215
L G IP + G L L+ L + LTG I I L KLK L ++ N SG IPD
Sbjct: 255 LSGPIPPSVGDLPYLEYLDFHKLPKLTGPIQPAIAKLTKLKSLSITWTNVSGPIPDFLAQ 434
Query: 216 LSDLLILDLSRNSLSGTLPVTLGRLISVLKLDLSHNFLEGKLLNEFGNLKNL-TLMDLRN 274
L +L L LS N+LSG +P +L RL ++L L+L N L G + FG K + L +
Sbjct: 435 LKNLDSLYLSFNNLSGHIPASLSRLPNLLSLNLDRNKLTGPIPASFGFFKKPGPNLFLSH 614
Query: 275 NRLCCGLVLSLQEMNSLEEMVLSNNPLGGDIRTLKWENLQNLVILELSNMELIGEIPESL 334
N+L + SL +++ E + S N L G+ L N + +L+LS L ++ +
Sbjct: 615 NQLSGPIPSSLGQLDP-ERIDFSRNKLTGNASLLFAHN-KKTQLLDLSRNLLSFDL-SKV 785
Query: 335 SQLKKLRFLGLSDNNITGNLSPKLETLPSLNALYLSGNNLKGEIQFSKGFFGKLGR--RF 392
K L +L L+ N I G++ L + +L +S N L G+I G+LG+ +F
Sbjct: 786 EFPKSLTWLDLNHNKIHGSIPVGLSAVQNLQQFNVSYNLLSGKIPQG----GELGKFDKF 953
Query: 393 GAWSNPKLC 401
+ N LC
Sbjct: 954 SYFHNKNLC 980
>TC18437 weakly similar to UP|Q9LVB3 (Q9LVB3) Receptor-like protein kinase,
partial (9%)
Length = 503
Score = 85.9 bits (211), Expect = 1e-17
Identities = 59/165 (35%), Positives = 89/165 (53%), Gaps = 1/165 (0%)
Frame = +3
Query: 168 LKNLQSLVLLENGLTGNIPQEIGNLVKLKRLVLSGNNFSGNIPDIFGGLSDLLILDLSRN 227
LKNL +N L+GNIP +GNL +L L L N G+IP + L L + N
Sbjct: 6 LKNLVRWAWQQNKLSGNIPTVMGNLTRLSELYLHTNRLQGSIPSTLRYCTKLQSLGAADN 185
Query: 228 SLSGTLPV-TLGRLISVLKLDLSHNFLEGKLLNEFGNLKNLTLMDLRNNRLCCGLVLSLQ 286
+++G++P TLG L ++ LD S N L G L +E GNLK+L+L+ L N+ + + L
Sbjct: 186 NITGSIPYQTLGYLEGLVNLDWSSNSLIGPLPSELGNLKHLSLLYLHANKWSGEIPMELG 365
Query: 287 EMNSLEEMVLSNNPLGGDIRTLKWENLQNLVILELSNMELIGEIP 331
+L E++L N G I + + ++L IL+LS+ IP
Sbjct: 366 AG*ALTELMLGRNLFRGSIPSF-LGSFRSLEILDLSSNNFTSMIP 497
Score = 77.0 bits (188), Expect = 6e-15
Identities = 54/151 (35%), Positives = 79/151 (51%), Gaps = 3/151 (1%)
Frame = +3
Query: 128 NKLPVSIPT--GNWEKLAESLESIEFRSNPGLIGNIPSTFGVLKNLQSLVLLENGLTGNI 185
NKL +IPT GN +L+E + L G+IPST LQSL +N +TG+I
Sbjct: 39 NKLSGNIPTVMGNLTRLSEL-----YLHTNRLQGSIPSTLRYCTKLQSLGAADNNITGSI 203
Query: 186 P-QEIGNLVKLKRLVLSGNNFSGNIPDIFGGLSDLLILDLSRNSLSGTLPVTLGRLISVL 244
P Q +G L L L S N+ G +P G L L +L L N SG +P+ LG ++
Sbjct: 204 PYQTLGYLEGLVNLDWSSNSLIGPLPSELGNLKHLSLLYLHANKWSGEIPMELGAG*ALT 383
Query: 245 KLDLSHNFLEGKLLNEFGNLKNLTLMDLRNN 275
+L L N G + + G+ ++L ++DL +N
Sbjct: 384 ELMLGRNLFRGSIPSFLGSFRSLEILDLSSN 476
Score = 63.9 bits (154), Expect = 5e-11
Identities = 43/107 (40%), Positives = 57/107 (53%)
Frame = +3
Query: 128 NKLPVSIPTGNWEKLAESLESIEFRSNPGLIGNIPSTFGVLKNLQSLVLLENGLTGNIPQ 187
N + SIP L E L ++++ SN LIG +PS G LK+L L L N +G IP
Sbjct: 183 NNITGSIPYQTLGYL-EGLVNLDWSSN-SLIGPLPSELGNLKHLSLLYLHANKWSGEIPM 356
Query: 188 EIGNLVKLKRLVLSGNNFSGNIPDIFGGLSDLLILDLSRNSLSGTLP 234
E+G L L+L N F G+IP G L ILDLS N+ + +P
Sbjct: 357 ELGAG*ALTELMLGRNLFRGSIPSFLGSFRSLEILDLSSNNFTSMIP 497
Score = 58.2 bits (139), Expect = 3e-09
Identities = 44/150 (29%), Positives = 69/150 (45%)
Frame = +3
Query: 239 RLISVLKLDLSHNFLEGKLLNEFGNLKNLTLMDLRNNRLCCGLVLSLQEMNSLEEMVLSN 298
+L ++++ N L G + GNL L+ + L NRL + +L+ L+ + ++
Sbjct: 3 KLKNLVRWAWQQNKLSGNIPTVMGNLTRLSELYLHTNRLQGSIPSTLRYCTKLQSLGAAD 182
Query: 299 NPLGGDIRTLKWENLQNLVILELSNMELIGEIPESLSQLKKLRFLGLSDNNITGNLSPKL 358
N + G I L+ LV L+ S+ LIG +P L LK L L L N +G + +L
Sbjct: 183 NNITGSIPYQTLGYLEGLVNLDWSSNSLIGPLPSELGNLKHLSLLYLHANKWSGEIPMEL 362
Query: 359 ETLPSLNALYLSGNNLKGEIQFSKGFFGKL 388
+L L L N +G I G F L
Sbjct: 363 GAG*ALTELMLGRNLFRGSIPSFLGSFRSL 452
Score = 30.0 bits (66), Expect = 0.86
Identities = 20/72 (27%), Positives = 33/72 (45%)
Frame = +3
Query: 336 QLKKLRFLGLSDNNITGNLSPKLETLPSLNALYLSGNNLKGEIQFSKGFFGKLGRRFGAW 395
+LK L N ++GN+ + L L+ LYL N L+G I + + KL A
Sbjct: 3 KLKNLVRWAWQQNKLSGNIPTVMGNLTRLSELYLHTNRLQGSIPSTLRYCTKLQSLGAAD 182
Query: 396 SNPKLCYPFELM 407
+N P++ +
Sbjct: 183 NNITGSIPYQTL 218
>BP043329
Length = 484
Score = 84.0 bits (206), Expect = 5e-17
Identities = 57/150 (38%), Positives = 80/150 (53%)
Frame = +3
Query: 145 SLESIEFRSNPGLIGNIPSTFGVLKNLQSLVLLENGLTGNIPQEIGNLVKLKRLVLSGNN 204
+L+ + NP G IP G L NL+ + L + L G IP IGNL KLK L L+ N+
Sbjct: 33 TLKMLNLSYNPFYPGRIPPEIGNLTNLEVVWLTQCNLVGVIPDSIGNLKKLKDLDLALND 212
Query: 205 FSGNIPDIFGGLSDLLILDLSRNSLSGTLPVTLGRLISVLKLDLSHNFLEGKLLNEFGNL 264
G+IP GL+ L ++L NSLSG LP +G L + LD S N L G++ E +L
Sbjct: 213 LYGSIPSSLTGLTSLRQIELYNNSLSGELPRGMGNLTELRLLDASMNHLTGRIPEELCSL 392
Query: 265 KNLTLMDLRNNRLCCGLVLSLQEMNSLEEM 294
L ++L NR L S+ + +L E+
Sbjct: 393 P-LESLNLYENRFEGELPASIADSPNLYEL 479
Score = 73.9 bits (180), Expect = 5e-14
Identities = 53/151 (35%), Positives = 84/151 (55%), Gaps = 1/151 (0%)
Frame = +3
Query: 232 TLPVTLGRLISVLKLDLSHN-FLEGKLLNEFGNLKNLTLMDLRNNRLCCGLVLSLQEMNS 290
T+P +LG L ++ L+LS+N F G++ E GNL NL ++ L L + S+ +
Sbjct: 3 TIPPSLGTLTTLKMLNLSYNPFYPGRIPPEIGNLTNLEVVWLTQCNLVGVIPDSIGNLKK 182
Query: 291 LEEMVLSNNPLGGDIRTLKWENLQNLVILELSNMELIGEIPESLSQLKKLRFLGLSDNNI 350
L+++ L+ N L G I + L +L +EL N L GE+P + L +LR L S N++
Sbjct: 183 LKDLDLALNDLYGSIPS-SLTGLTSLRQIELYNNSLSGELPRGMGNLTELRLLDASMNHL 359
Query: 351 TGNLSPKLETLPSLNALYLSGNNLKGEIQFS 381
TG + +L +LP L +L L N +GE+ S
Sbjct: 360 TGRIPEELCSLP-LESLNLYENRFEGELPAS 449
Score = 56.2 bits (134), Expect = 1e-08
Identities = 35/100 (35%), Positives = 55/100 (55%)
Frame = +3
Query: 284 SLQEMNSLEEMVLSNNPLGGDIRTLKWENLQNLVILELSNMELIGEIPESLSQLKKLRFL 343
SL + +L+ + LS NP + NL NL ++ L+ L+G IP+S+ LKKL+ L
Sbjct: 15 SLGTLTTLKMLNLSYNPFYPGRIPPEIGNLTNLEVVWLTQCNLVGVIPDSIGNLKKLKDL 194
Query: 344 GLSDNNITGNLSPKLETLPSLNALYLSGNNLKGEIQFSKG 383
L+ N++ G++ L L SL + L N+L GE+ G
Sbjct: 195 DLALNDLYGSIPSSLTGLTSLRQIELYNNSLSGELPRGMG 314
>TC19434 similar to UP|Q94F63 (Q94F63) Somatic embryogenesis receptor-like
kinase 2, partial (26%)
Length = 550
Score = 82.8 bits (203), Expect = 1e-16
Identities = 49/110 (44%), Positives = 65/110 (58%)
Frame = +2
Query: 166 GVLKNLQSLVLLENGLTGNIPQEIGNLVKLKRLVLSGNNFSGNIPDIFGGLSDLLILDLS 225
G L NLQ L L N +TG IP E+GNL L L L NN +GNIP G L L L L+
Sbjct: 35 GQLSNLQYLELYSNNMTGKIPDELGNLTNLVSLDLYLNNLTGNIPSTLGNLGKLRFLRLN 214
Query: 226 RNSLSGTLPVTLGRLISVLKLDLSHNFLEGKLLNEFGNLKNLTLMDLRNN 275
N+LSG +P+TL + S+ LDLS+N L+G ++ G+ T + +NN
Sbjct: 215 NNTLSGGIPMTLTNVSSLQVLDLSNNQLKG-VVPVNGSFSLFTPISYQNN 361
Score = 76.6 bits (187), Expect = 8e-15
Identities = 44/91 (48%), Positives = 54/91 (58%)
Frame = +2
Query: 145 SLESIEFRSNPGLIGNIPSTFGVLKNLQSLVLLENGLTGNIPQEIGNLVKLKRLVLSGNN 204
+L+ +E SN + G IP G L NL SL L N LTGNIP +GNL KL+ L L+ N
Sbjct: 47 NLQYLELYSN-NMTGKIPDELGNLTNLVSLDLYLNNLTGNIPSTLGNLGKLRFLRLNNNT 223
Query: 205 FSGNIPDIFGGLSDLLILDLSRNSLSGTLPV 235
SG IP +S L +LDLS N L G +PV
Sbjct: 224 LSGGIPMTLTNVSSLQVLDLSNNQLKGVVPV 316
Score = 68.9 bits (167), Expect = 2e-12
Identities = 37/90 (41%), Positives = 56/90 (62%)
Frame = +2
Query: 188 EIGNLVKLKRLVLSGNNFSGNIPDIFGGLSDLLILDLSRNSLSGTLPVTLGRLISVLKLD 247
++G L L+ L L NN +G IPD G L++L+ LDL N+L+G +P TLG L + L
Sbjct: 29 QLGQLSNLQYLELYSNNMTGKIPDELGNLTNLVSLDLYLNNLTGNIPSTLGNLGKLRFLR 208
Query: 248 LSHNFLEGKLLNEFGNLKNLTLMDLRNNRL 277
L++N L G + N+ +L ++DL NN+L
Sbjct: 209 LNNNTLSGGIPMTLTNVSSLQVLDLSNNQL 298
Score = 65.1 bits (157), Expect = 2e-11
Identities = 36/90 (40%), Positives = 55/90 (61%)
Frame = +2
Query: 214 GGLSDLLILDLSRNSLSGTLPVTLGRLISVLKLDLSHNFLEGKLLNEFGNLKNLTLMDLR 273
G LS+L L+L N+++G +P LG L +++ LDL N L G + + GNL L + L
Sbjct: 35 GQLSNLQYLELYSNNMTGKIPDELGNLTNLVSLDLYLNNLTGNIPSTLGNLGKLRFLRLN 214
Query: 274 NNRLCCGLVLSLQEMNSLEEMVLSNNPLGG 303
NN L G+ ++L ++SL+ + LSNN L G
Sbjct: 215 NNTLSGGIPMTLTNVSSLQVLDLSNNQLKG 304
Score = 62.0 bits (149), Expect = 2e-10
Identities = 43/119 (36%), Positives = 63/119 (52%)
Frame = +2
Query: 285 LQEMNSLEEMVLSNNPLGGDIRTLKWENLQNLVILELSNMELIGEIPESLSQLKKLRFLG 344
L ++++L+ + L +N + G I + NL NLV L+L L G IP +L L KLRFL
Sbjct: 32 LGQLSNLQYLELYSNNMTGKIPD-ELGNLTNLVSLDLYLNNLTGNIPSTLGNLGKLRFLR 208
Query: 345 LSDNNITGNLSPKLETLPSLNALYLSGNNLKGEIQFSKGFFGKLGRRFGAWSNPKLCYP 403
L++N ++G + L + SL L LS N LKG + + F L +NP L P
Sbjct: 209 LNNNTLSGGIPMTLTNVSSLQVLDLSNNQLKGVVPVNGSF--SLFTPISYQNNPGLIQP 379
Score = 59.3 bits (142), Expect = 1e-09
Identities = 42/121 (34%), Positives = 65/121 (53%)
Frame = +2
Query: 237 LGRLISVLKLDLSHNFLEGKLLNEFGNLKNLTLMDLRNNRLCCGLVLSLQEMNSLEEMVL 296
LG+L ++ L+L N + GK+ +E GNL NL +DL N L + +L + L + L
Sbjct: 32 LGQLSNLQYLELYSNNMTGKIPDELGNLTNLVSLDLYLNNLTGNIPSTLGNLGKLRFLRL 211
Query: 297 SNNPLGGDIRTLKWENLQNLVILELSNMELIGEIPESLSQLKKLRFLGLSDNNITGNLSP 356
+NN L G I + N+ +L +L+LSN +L G +P + S F +S N G + P
Sbjct: 212 NNNTLSGGI-PMTLTNVSSLQVLDLSNNQLKGVVPVNGS---FSLFTPISYQNNPGLIQP 379
Query: 357 K 357
K
Sbjct: 380 K 382
>TC14470 similar to UP|AAQ56728 (AAQ56728) Polygalacturonase inhibiting
protein, partial (93%)
Length = 1195
Score = 82.0 bits (201), Expect = 2e-16
Identities = 76/230 (33%), Positives = 112/230 (48%), Gaps = 10/230 (4%)
Frame = +3
Query: 159 GNIPSTFGVLKNLQSLVLLE-NGLTGNIPQEIGNLVKLKRLVLSGNNFSGNIPDIFGGLS 217
G IP + G L LQ L + LTG I I L L+ L+L+ N SG IPD L
Sbjct: 249 GQIPPSVGDLPYLQVLEFHKLPKLTGPIQPAIAKLTNLRWLILTWTNISGPIPDFLSELP 428
Query: 218 DLLILDLSRNSLSGTLPVTLGRLISVLKLDLSHNFLEGKLLNEFGNLKNLTL-MDLRNNR 276
+L L LS N+L+G +P +L +L +++ L L N L G + + FG+ K + + L +N+
Sbjct: 429 NLQWLHLSFNNLTGPIPSSLSKLPNLIDLRLDRNRLTGPIPDSFGSFKKPGIDLTLSHNQ 608
Query: 277 LCCGLVLSLQEMNSLEEMVLSNNPLGGDIRTLKWEN--------LQNLVILELSNMELIG 328
L + SL ++ + S N L GD L N +NL+ ++S +EL
Sbjct: 609 LSGPIPTSLGLIDP-NRIDFSRNMLEGDASMLFGVNKTTEMIDLSRNLLTFDMSKVEL-- 779
Query: 329 EIPESLSQLKKLRFLGLSDNNITGNLSPKLETLPSLNALYLSGNNLKGEI 378
P SL L L+ N I G++ +L + L A +S N L GEI
Sbjct: 780 --PTSLIS------LDLNHNQIYGSIPQELIKVDFLQAFNVSYNRLCGEI 905
Score = 60.5 bits (145), Expect = 6e-10
Identities = 60/178 (33%), Positives = 79/178 (43%), Gaps = 2/178 (1%)
Frame = +3
Query: 157 LIGNIPSTFGVLKNLQSLVLLENGLTGNIPQEIGNLVKLKRLVLSGNNFSGNIPDIFGGL 216
L G IPS+ L NL L L N LTG IP G+ K P I
Sbjct: 462 LTGPIPSSLSKLPNLIDLRLDRNRLTGPIPDSFGSFKK---------------PGI---- 584
Query: 217 SDLLILDLSRNSLSGTLPVTLGRLISVLKLDLSHNFLEGKLLNEFGNLKNLTLMDLRNNR 276
L LS N LSG +P +LG LI ++D S N LEG FG K ++DL N
Sbjct: 585 ----DLTLSHNQLSGPIPTSLG-LIDPNRIDFSRNMLEGDASMLFGVNKTTEMIDLSRNL 749
Query: 277 LCCGLVLSLQEMNSLEEMVLSNNPLGGDI--RTLKWENLQNLVILELSNMELIGEIPE 332
L + ++ SL + L++N + G I +K + LQ +S L GEIP+
Sbjct: 750 LTFDM-SKVELPTSLISLDLNHNQIYGSIPQELIKVDFLQ---AFNVSYNRLCGEIPQ 911
Score = 52.4 bits (124), Expect = 2e-07
Identities = 38/107 (35%), Positives = 54/107 (49%)
Frame = +3
Query: 283 LSLQEMNSLEEMVLSNNPLGGDIRTLKWENLQNLVILELSNMELIGEIPESLSQLKKLRF 342
L + E + L ++ P + L+W +IL +N+ G IP+ LS+L L++
Sbjct: 285 LQVLEFHKLPKLTGPIQPAIAKLTNLRW------LILTWTNIS--GPIPDFLSELPNLQW 440
Query: 343 LGLSDNNITGNLSPKLETLPSLNALYLSGNNLKGEIQFSKGFFGKLG 389
L LS NN+TG + L LP+L L L N L G I S G F K G
Sbjct: 441 LHLSFNNLTGPIPSSLSKLPNLIDLRLDRNRLTGPIPDSFGSFKKPG 581
>TC14398 similar to UP|Q96477 (Q96477) LRR protein, partial (86%)
Length = 982
Score = 77.8 bits (190), Expect = 4e-15
Identities = 39/98 (39%), Positives = 61/98 (61%)
Frame = +1
Query: 181 LTGNIPQEIGNLVKLKRLVLSGNNFSGNIPDIFGGLSDLLILDLSRNSLSGTLPVTLGRL 240
L+G++ ++GNL L+ L L NN G IP+ G L L+ LDL N++SG++P +LG L
Sbjct: 358 LSGHLVPDLGNLHSLQYLELYENNIQGTIPEELGNLQSLISLDLYHNNVSGSIPSSLGNL 537
Query: 241 ISVLKLDLSHNFLEGKLLNEFGNLKNLTLMDLRNNRLC 278
++ L L++N L G++ L NL ++D+ NN LC
Sbjct: 538 KNLRFLRLNNNHLTGQIPKSLSTLPNLKVLDVSNNNLC 651
Score = 77.8 bits (190), Expect = 4e-15
Identities = 51/138 (36%), Positives = 74/138 (52%), Gaps = 6/138 (4%)
Frame = +1
Query: 197 RLVLSGNNFSGNIPDIFGGLSDLLILDLSRNSLSGTLPVTLGRLISVLKLDLSHNFLEGK 256
R+ L N SG++ G L L L+L N++ GT+P LG L S++ LDL HN + G
Sbjct: 334 RVDLGNLNLSGHLVPDLGNLHSLQYLELYENNIQGTIPEELGNLQSLISLDLYHNNVSGS 513
Query: 257 LLNEFGNLKNLTLMDLRNNRLCCGLVLSLQEMNSLEEMVLSNNPLGGDIRT------LKW 310
+ + GNLKNL + L NN L + SL + +L+ + +SNN L G I T +
Sbjct: 514 IPSSLGNLKNLRFLRLNNNHLTGQIPKSLSTLPNLKVLDVSNNNLCGPIPTSGPFEHIPL 693
Query: 311 ENLQNLVILELSNMELIG 328
+N +N LE EL+G
Sbjct: 694 DNFENNPRLE--GPELLG 741
Score = 77.8 bits (190), Expect = 4e-15
Identities = 49/119 (41%), Positives = 70/119 (58%)
Frame = +1
Query: 266 NLTLMDLRNNRLCCGLVLSLQEMNSLEEMVLSNNPLGGDIRTLKWENLQNLVILELSNME 325
++T +DL N L LV L ++SL+ + L N + G I + NLQ+L+ L+L +
Sbjct: 325 SVTRVDLGNLNLSGHLVPDLGNLHSLQYLELYENNIQGTIPE-ELGNLQSLISLDLYHNN 501
Query: 326 LIGEIPESLSQLKKLRFLGLSDNNITGNLSPKLETLPSLNALYLSGNNLKGEIQFSKGF 384
+ G IP SL LK LRFL L++N++TG + L TLP+L L +S NNL G I S F
Sbjct: 502 VSGSIPSSLGNLKNLRFLRLNNNHLTGQIPKSLSTLPNLKVLDVSNNNLCGPIPTSGPF 678
Score = 77.8 bits (190), Expect = 4e-15
Identities = 48/122 (39%), Positives = 69/122 (56%)
Frame = +1
Query: 154 NPGLIGNIPSTFGVLKNLQSLVLLENGLTGNIPQEIGNLVKLKRLVLSGNNFSGNIPDIF 213
N L G++ G L +LQ L L EN + G IP+E+GNL L L L NN SG+IP
Sbjct: 349 NLNLSGHLVPDLGNLHSLQYLELYENNIQGTIPEELGNLQSLISLDLYHNNVSGSIPSSL 528
Query: 214 GGLSDLLILDLSRNSLSGTLPVTLGRLISVLKLDLSHNFLEGKLLNEFGNLKNLTLMDLR 273
G L +L L L+ N L+G +P +L L ++ LD+S+N L G + G +++ L +
Sbjct: 529 GNLKNLRFLRLNNNHLTGQIPKSLSTLPNLKVLDVSNNNLCGPIPTS-GPFEHIPLDNFE 705
Query: 274 NN 275
NN
Sbjct: 706 NN 711
>TC14712 similar to UP|PGI2_PHAVU (P58822) Polygalacturonase inhibitor 2
precursor (Polygalacturonase-inhibiting protein)
(PGIP-2), partial (51%)
Length = 824
Score = 77.0 bits (188), Expect = 6e-15
Identities = 55/175 (31%), Positives = 88/175 (49%), Gaps = 1/175 (0%)
Frame = +1
Query: 181 LTGNIPQEIGNLVKLKRLVLSGNNFSGNIPDIFGGLSDLLI-LDLSRNSLSGTLPVTLGR 239
LTG +P I L L + + N SG IPD +G S+L L L+RN LSG +P +L +
Sbjct: 16 LTGPLPSSISTLPNLVGITANDNKLSGAIPDSYGSFSNLFTSLTLNRNQLSGKIPASLSK 195
Query: 240 LISVLKLDLSHNFLEGKLLNEFGNLKNLTLMDLRNNRLCCGLVLSLQEMNSLEEMVLSNN 299
L ++ +DLS N LEG FG+ KN ++++L+ N
Sbjct: 196 L-NLAFVDLSMNMLEGDASVFFGSKKN------------------------TQKIILARN 300
Query: 300 PLGGDIRTLKWENLQNLVILELSNMELIGEIPESLSQLKKLRFLGLSDNNITGNL 354
L D+ + + NL ++L N + G++P+ L+ LK L+ L +S N++ G +
Sbjct: 301 SLAFDLGKVGLSS--NLNTIDLRNNRVYGKLPQELTGLKFLKKLNVSYNSLCGQI 459
Score = 28.9 bits (63), Expect = 1.9
Identities = 15/39 (38%), Positives = 20/39 (50%)
Frame = +1
Query: 350 ITGNLSPKLETLPSLNALYLSGNNLKGEIQFSKGFFGKL 388
+TG L + TLP+L + + N L G I S G F L
Sbjct: 16 LTGPLPSSISTLPNLVGITANDNKLSGAIPDSYGSFSNL 132
>TC11433 similar to UP|AAR99871 (AAR99871) Strubbelig receptor family 3,
partial (24%)
Length = 565
Score = 72.8 bits (177), Expect = 1e-13
Identities = 47/120 (39%), Positives = 68/120 (56%)
Frame = +2
Query: 153 SNPGLIGNIPSTFGVLKNLQSLVLLENGLTGNIPQEIGNLVKLKRLVLSGNNFSGNIPDI 212
SN + G+IPS+ V +++ L N TG+IP + L L + L+ N+ SG IPD
Sbjct: 137 SNNNIGGSIPSSLPV--TMRNFFLAANQFTGSIPTSLSTLTGLTDMSLNENHLSGEIPDA 310
Query: 213 FGGLSDLLILDLSRNSLSGTLPVTLGRLISVLKLDLSHNFLEGKLLNEFGNLKNLTLMDL 272
F L+ L+ LDLS N+LSG LP ++ L ++ L L +N L G L L++L L DL
Sbjct: 311 FQSLTQLINLDLSTNNLSGELPPSVENLSALTTLRLQNNQLSGTL----DVLQDLPLQDL 478
Score = 70.9 bits (172), Expect = 4e-13
Identities = 50/150 (33%), Positives = 79/150 (52%)
Frame = +2
Query: 156 GLIGNIPSTFGVLKNLQSLVLLENGLTGNIPQEIGNLVKLKRLVLSGNNFSGNIPDIFGG 215
G +G+ STF + + L N + G+IP + V ++ L+ N F+G+IP
Sbjct: 83 GELGDNLSTFVTISVID---LSNNNIGGSIPSSLP--VTMRNFFLAANQFTGSIPTSLST 247
Query: 216 LSDLLILDLSRNSLSGTLPVTLGRLISVLKLDLSHNFLEGKLLNEFGNLKNLTLMDLRNN 275
L+ L + L+ N LSG +P L ++ LDLS N L G+L NL LT + L+NN
Sbjct: 248 LTGLTDMSLNENHLSGEIPDAFQSLTQLINLDLSTNNLSGELPPSVENLSALTTLRLQNN 427
Query: 276 RLCCGLVLSLQEMNSLEEMVLSNNPLGGDI 305
+L G + LQ++ L+++ + NN G I
Sbjct: 428 QL-SGTLDVLQDL-PLQDLNVENNQFAGPI 511
Score = 60.8 bits (146), Expect = 5e-10
Identities = 42/136 (30%), Positives = 66/136 (47%)
Frame = +2
Query: 243 VLKLDLSHNFLEGKLLNEFGNLKNLTLMDLRNNRLCCGLVLSLQEMNSLEEMVLSNNPLG 302
+ K+ L+ L G+L + ++++DL NN + + SL ++ L+ N
Sbjct: 47 IQKIVLNGANLGGELGDNLSTFVTISVIDLSNNNIGGSIPSSLPV--TMRNFFLAANQFT 220
Query: 303 GDIRTLKWENLQNLVILELSNMELIGEIPESLSQLKKLRFLGLSDNNITGNLSPKLETLP 362
G I T L L + L+ L GEIP++ L +L L LS NN++G L P +E L
Sbjct: 221 GSIPT-SLSTLTGLTDMSLNENHLSGEIPDAFQSLTQLINLDLSTNNLSGELPPSVENLS 397
Query: 363 SLNALYLSGNNLKGEI 378
+L L L N L G +
Sbjct: 398 ALTTLRLQNNQLSGTL 445
>BP053628
Length = 567
Score = 71.2 bits (173), Expect = 3e-13
Identities = 53/178 (29%), Positives = 89/178 (49%), Gaps = 19/178 (10%)
Frame = +1
Query: 207 GNIPDIFGGLSDLLILDLSRNSLSGTLPVTLGRLISVLKLDLSHNFLEGKLLNEFGNLKN 266
G++P L L LDLS N L G++P + + ++ L L NFL G + F + N
Sbjct: 31 GSLPSSIHRLYALEALDLSSNYLYGSIPPKIFTIGNLQTLRLGDNFLNGTIPTLFNSSSN 210
Query: 267 LTLMDLRNNRLCCGLVLSLQEMNSLEEMVLSNNPLGGD------IRTLKWENLQ------ 314
L ++ L+NNRL S+ + +L E+ +S N + G +R+L+ +L+
Sbjct: 211 LKVLSLKNNRLTGQFPSSILSITTLIEIDMSTNEISGSLQDFTGLRSLEQLDLRENRLDS 390
Query: 315 -------NLVILELSNMELIGEIPESLSQLKKLRFLGLSDNNITGNLSPKLETLPSLN 365
L+ + LS G+IP+ QLK L+ L +S N +TG +L +LP+++
Sbjct: 391 ALPKMPPRLMGIFLSRNSFSGQIPKHYGQLKMLQQLDVSFNALTGTXPAELFSLPNIS 564
>TC9224 similar to GB|AAP21230.1|30102624|BT006422 At3g49750 {Arabidopsis
thaliana;}, partial (77%)
Length = 1318
Score = 70.5 bits (171), Expect = 6e-13
Identities = 53/160 (33%), Positives = 72/160 (44%), Gaps = 4/160 (2%)
Frame = +1
Query: 153 SNPGLIGNIPSTFGVLKNLQSLVLLENGLTGNIPQEIGNLVKLKRLVLSGNNFSGNIPDI 212
+N L G I NLQ+L L N LTG IP ++ +LV L L LS N G IP
Sbjct: 577 NNLALHGTISPFLANCTNLQALDLSSNFLTGPIPPDLQSLVNLAVLNLSANRLEGEIPPQ 756
Query: 213 FGGLSDLLILDLSRNSLSGTLPVTLGRLISVLKLDLSHNFLEG----KLLNEFGNLKNLT 268
+ L I+DL +N L+G +P LG L+ + D+S+N L G L N GNL
Sbjct: 757 LTMCAYLNIIDLHQNLLTGPIPQQLGLLVRLSAFDVSNNRLAGPIPSSLTNRSGNLPRFN 936
Query: 269 LMDLRNNRLCCGLVLSLQEMNSLEEMVLSNNPLGGDIRTL 308
N+ G L L + + LG + +L
Sbjct: 937 ASSFVGNKDLYGYPLPPLRSKGLSILAIVGIGLGSGLASL 1056
Score = 57.0 bits (136), Expect = 7e-09
Identities = 52/174 (29%), Positives = 76/174 (42%)
Frame = +1
Query: 179 NGLTGNIPQEIGNLVKLKRLVLSGNNFSGNIPDIFGGLSDLLILDLSRNSLSGTLPVTLG 238
N T N+ I N ++ +L L+ G I ++L LDLS N L+G +P L
Sbjct: 511 NDSTSNLQGAICNNGRIYKLSLNNLALHGTISPFLANCTNLQALDLSSNFLTGPIPPDLQ 690
Query: 239 RLISVLKLDLSHNFLEGKLLNEFGNLKNLTLMDLRNNRLCCGLVLSLQEMNSLEEMVLSN 298
L+++ L+LS N LEG++ + L ++DL N L + L + L +SN
Sbjct: 691 SLVNLAVLNLSANRLEGEIPPQLTMCAYLNIIDLHQNLLTGPIPQQLGLLVRLSAFDVSN 870
Query: 299 NPLGGDIRTLKWENLQNLVILELSNMELIGEIPESLSQLKKLRFLGLSDNNITG 352
N L G I + NL S+ +G L LR GLS I G
Sbjct: 871 NRLAGPIPSSLTNRSGNLPRFNASS--FVGNKDLYGYPLPPLRSKGLSILAIVG 1026
Score = 50.4 bits (119), Expect = 6e-07
Identities = 44/130 (33%), Positives = 63/130 (47%), Gaps = 1/130 (0%)
Frame = +1
Query: 253 LEGKLLNEFGNLKNLTLMDLRNNRLCCGLVLS-LQEMNSLEEMVLSNNPLGGDIRTLKWE 311
L+G + N G + L+L NN G + L +L+ + LS+N L G I +
Sbjct: 529 LQGAICNN-GRIYKLSL----NNLALHGTISPFLANCTNLQALDLSSNFLTGPIPP-DLQ 690
Query: 312 NLQNLVILELSNMELIGEIPESLSQLKKLRFLGLSDNNITGNLSPKLETLPSLNALYLSG 371
+L NL +L LS L GEIP L+ L + L N +TG + +L L L+A +S
Sbjct: 691 SLVNLAVLNLSANRLEGEIPPQLTMCAYLNIIDLHQNLLTGPIPQQLGLLVRLSAFDVSN 870
Query: 372 NNLKGEIQFS 381
N L G I S
Sbjct: 871 NRLAGPIPSS 900
>TC15223 similar to UP|CAE76632 (CAE76632) Leucine rich repeat protein
precursor, partial (35%)
Length = 876
Score = 68.6 bits (166), Expect = 2e-12
Identities = 42/117 (35%), Positives = 62/117 (52%)
Frame = +3
Query: 183 GNIPQEIGNLVKLKRLVLSGNNFSGNIPDIFGGLSDLLILDLSRNSLSGTLPVTLGRLIS 242
G++P E G + L L L N+ SG IP S + IL+LSRN L GT+P G
Sbjct: 3 GSVPAEFGRMRVLSTLNLDSNSLSGQIPSTLLSNSGMGILNLSRNGLEGTIPDVFGPNSY 182
Query: 243 VLKLDLSHNFLEGKLLNEFGNLKNLTLMDLRNNRLCCGLVLSLQEMNSLEEMVLSNN 299
+ LDLS+N L+G++ + K + +DL +N L CG + + + LE S+N
Sbjct: 183 FMALDLSYNGLQGRVPGSLCSAKYIGHLDLSHNHL-CGSIPMGEPFDHLEASSFSSN 350
Score = 68.2 bits (165), Expect = 3e-12
Identities = 40/99 (40%), Positives = 56/99 (56%)
Frame = +3
Query: 159 GNIPSTFGVLKNLQSLVLLENGLTGNIPQEIGNLVKLKRLVLSGNNFSGNIPDIFGGLSD 218
G++P+ FG ++ L +L L N L+G IP + + + L LS N G IPD+FG S
Sbjct: 3 GSVPAEFGRMRVLSTLNLDSNSLSGQIPSTLLSNSGMGILNLSRNGLEGTIPDVFGPNSY 182
Query: 219 LLILDLSRNSLSGTLPVTLGRLISVLKLDLSHNFLEGKL 257
+ LDLS N L G +P +L + LDLSHN L G +
Sbjct: 183 FMALDLSYNGLQGRVPGSLCSAKYIGHLDLSHNHLCGSI 299
>BI418931
Length = 516
Score = 68.6 bits (166), Expect = 2e-12
Identities = 48/134 (35%), Positives = 68/134 (49%), Gaps = 9/134 (6%)
Frame = +2
Query: 181 LTGNIPQEIGNLVKLKRLVLSGNNFSGNIPDIFGGLSDLLI---------LDLSRNSLSG 231
L+G IP + L RL LSGN F+G + G S+LL+ LD+S NSL G
Sbjct: 17 LSGTIPSGLVTSSSLARLNLSGNQFTGPLQLQGSGSSELLLMSPSQHMEYLDVSNNSLEG 196
Query: 232 TLPVTLGRLISVLKLDLSHNFLEGKLLNEFGNLKNLTLMDLRNNRLCCGLVLSLQEMNSL 291
LP + ++ + L+L+ N G+L NE G L L +DL NN+ + L +SL
Sbjct: 197 VLPTEIDKMGGLRLLNLARNGFSGELPNELGKLVYLEYLDLSNNKFTGHIPDRLS--SSL 370
Query: 292 EEMVLSNNPLGGDI 305
+SNN L G +
Sbjct: 371 TAFNVSNNDLSGHV 412
Score = 60.5 bits (145), Expect = 6e-10
Identities = 35/85 (41%), Positives = 48/85 (56%)
Frame = +2
Query: 153 SNPGLIGNIPSTFGVLKNLQSLVLLENGLTGNIPQEIGNLVKLKRLVLSGNNFSGNIPDI 212
SN L G +P+ + L+ L L NG +G +P E+G LV L+ L LS N F+G+IPD
Sbjct: 176 SNNSLEGVLPTEIDKMGGLRLLNLARNGFSGELPNELGKLVYLEYLDLSNNKFTGHIPDR 355
Query: 213 FGGLSDLLILDLSRNSLSGTLPVTL 237
S L ++S N LSG +P L
Sbjct: 356 LS--SSLTAFNVSNNDLSGHVPKNL 424
Score = 50.8 bits (120), Expect = 5e-07
Identities = 43/143 (30%), Positives = 67/143 (46%), Gaps = 9/143 (6%)
Frame = +2
Query: 229 LSGTLPVTLGRLISVLKLDLSHNFLEGKLLNEFGNLKNLTLM---------DLRNNRLCC 279
LSGT+P L S+ +L+LS N G L + L LM D+ NN L
Sbjct: 17 LSGTIPSGLVTSSSLARLNLSGNQFTGPLQLQGSGSSELLLMSPSQHMEYLDVSNNSLEG 196
Query: 280 GLVLSLQEMNSLEEMVLSNNPLGGDIRTLKWENLQNLVILELSNMELIGEIPESLSQLKK 339
L + +M L + L+ N G++ + L L L+LSN + G IP+ LS
Sbjct: 197 VLPTEIDKMGGLRLLNLARNGFSGELPN-ELGKLVYLEYLDLSNNKFTGHIPDRLS--SS 367
Query: 340 LRFLGLSDNNITGNLSPKLETLP 362
L +S+N+++G++ L+ P
Sbjct: 368 LTAFNVSNNDLSGHVPKNLQHFP 436
Score = 49.3 bits (116), Expect = 1e-06
Identities = 31/87 (35%), Positives = 48/87 (54%)
Frame = +2
Query: 169 KNLQSLVLLENGLTGNIPQEIGNLVKLKRLVLSGNNFSGNIPDIFGGLSDLLILDLSRNS 228
++++ L + N L G +P EI + L+ L L+ N FSG +P+ G L L LDLS N
Sbjct: 152 QHMEYLDVSNNSLEGVLPTEIDKMGGLRLLNLARNGFSGELPNELGKLVYLEYLDLSNNK 331
Query: 229 LSGTLPVTLGRLISVLKLDLSHNFLEG 255
+G +P L S+ ++S+N L G
Sbjct: 332 FTGHIPDRLSS--SLTAFNVSNNDLSG 406
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.319 0.139 0.413
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,636,376
Number of Sequences: 28460
Number of extensions: 120193
Number of successful extensions: 1105
Number of sequences better than 10.0: 120
Number of HSP's better than 10.0 without gapping: 885
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1009
length of query: 475
length of database: 4,897,600
effective HSP length: 94
effective length of query: 381
effective length of database: 2,222,360
effective search space: 846719160
effective search space used: 846719160
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 57 (26.6 bits)
Medicago: description of AC146570.9


