

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146570.4 - phase: 0
(338 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
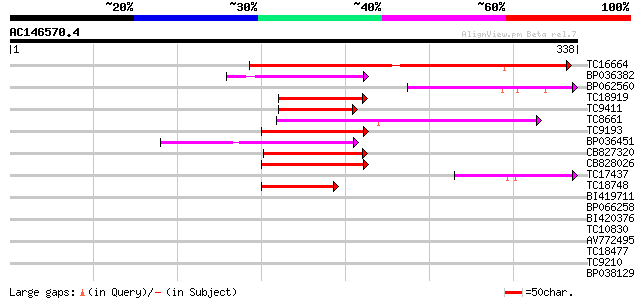
Score E
Sequences producing significant alignments: (bits) Value
TC16664 similar to UP|BAD09830 (BAD09830) DNA-binding protein-li... 135 7e-33
BP036382 58 2e-09
BP062560 55 2e-08
TC18919 similar to GB|AAN73298.1|25141207|BT002301 At4g16430/dl4... 55 2e-08
TC9411 similar to UP|Q41102 (Q41102) Phaseolin G-box binding pro... 53 8e-08
TC8661 weakly similar to UP|O81348 (O81348) Symbiotic ammonium t... 52 1e-07
TC9193 similar to GB|AAN28890.1|23506061|AY143951 At4g02590/T10P... 50 4e-07
BP036451 50 7e-07
CB827320 49 1e-06
CB828026 48 3e-06
TC17437 48 3e-06
TC18748 similar to GB|AAN28890.1|23506061|AY143951 At4g02590/T10... 41 3e-04
BI419711 38 0.002
BP066258 36 0.008
BI420376 36 0.011
TC10830 weakly similar to UP|Q94JL9 (Q94JL9) At1g68400/T2E12_5, ... 35 0.023
AV772495 33 0.089
TC18477 32 0.15
TC9210 32 0.15
BP038129 30 0.44
>TC16664 similar to UP|BAD09830 (BAD09830) DNA-binding protein-like, partial
(22%)
Length = 1067
Score = 135 bits (341), Expect = 7e-33
Identities = 75/199 (37%), Positives = 122/199 (60%), Gaps = 7/199 (3%)
Frame = +1
Query: 144 KMSAQEIMEAKALAASKSHSEAERRRRERINNHLAKLRSLLPSTTKTDKASLLAEVIQHV 203
++ + + +++ A ++HSEAERRRR RIN HL LR+++P K DKASLLAEVI H+
Sbjct: 238 QLERKGVSPERSIEALRNHSEAERRRRARINAHLDTLRTVIPGANKMDKASLLAEVITHL 417
Query: 204 KELKRQTSLIAETSPVPTECDELTVDAAADDEDYGSNGNKFIIKASLCCDDRSDLLPELI 263
KELK + +E +P + DE+ V ++++ G NG + I+ASLCC+ + LL ++
Sbjct: 418 KELKTNAAQASEGLMIPKDNDEIRV----EEQEGGLNGFPYSIRASLCCEYKPGLLSDIR 585
Query: 264 KTLKALRLRTLKADITTLGGRVKNVLFITG-------EEDDHEYCISSIQEALKAVMEKS 316
+ L AL L ++A+I TLGGR+KNV I + + ++ S+ +AL++V+++
Sbjct: 586 QALDALHLMIMRAEIATLGGRMKNVFVIISCKEQNFEDAEYRQFLAGSVHQALRSVLDRF 765
Query: 317 VGDESASGSVKRQRTNIIS 335
+ S KR+R +I S
Sbjct: 766 SVSQDILESRKRRRISIFS 822
>BP036382
Length = 511
Score = 58.2 bits (139), Expect = 2e-09
Identities = 35/85 (41%), Positives = 51/85 (59%)
Frame = +3
Query: 130 SGSGPFGGFQGEVGKMSAQEIMEAKALAASKSHSEAERRRRERINNHLAKLRSLLPSTTK 189
SGS GG Q + A+ + A+ A+ HS AER RRERI + L+ L+P+ K
Sbjct: 54 SGSAGGGGNQXQ-----AKPRVRARRGQATDPHSIAERLRRERIAERMKALQELVPNANK 218
Query: 190 TDKASLLAEVIQHVKELKRQTSLIA 214
TDKAS+L E+I +VK L+ Q +++
Sbjct: 219 TDKASMLDEIIDYVKFLQLQVKVLS 293
>BP062560
Length = 564
Score = 55.1 bits (131), Expect = 2e-08
Identities = 38/111 (34%), Positives = 62/111 (55%), Gaps = 10/111 (9%)
Frame = -1
Query: 238 GSNGNKFIIKASLCCDDRSDLLPELIKTLKALRLRTLKADITTLGGRVKNVLFIT---GE 294
G A++C D +S++L +L +TL AL+L+ +KA+++TLG R+KNV T E
Sbjct: 561 GEENESISYMATVCSDYQSEILSDLKQTLDALQLQLVKAELSTLGERMKNVFVFTCCXRE 382
Query: 295 EDDHEYC---ISSIQEALKAVMEKSVG----DESASGSVKRQRTNIISISN 338
+ E C S++ +AL +V+EK+ AS KR+R + I S+
Sbjct: 381 NVNIEACQALASNVHQALGSVLEKASNALEFSFRASDRCKRRRLSFIESSS 229
>TC18919 similar to GB|AAN73298.1|25141207|BT002301 At4g16430/dl4240w
{Arabidopsis thaliana;}, partial (29%)
Length = 604
Score = 54.7 bits (130), Expect = 2e-08
Identities = 23/53 (43%), Positives = 39/53 (73%)
Frame = +3
Query: 161 SHSEAERRRRERINNHLAKLRSLLPSTTKTDKASLLAEVIQHVKELKRQTSLI 213
+H EAER+RRE++N LR+++P+ +K DKASLL + I H+ +L+++ L+
Sbjct: 411 NHVEAERQRREKLNQRFYALRAVVPNISKMDKASLLGDAITHITDLQKKIKLL 569
>TC9411 similar to UP|Q41102 (Q41102) Phaseolin G-box binding protein PG2
(Fragment), partial (30%)
Length = 705
Score = 52.8 bits (125), Expect = 8e-08
Identities = 23/47 (48%), Positives = 35/47 (73%)
Frame = +2
Query: 161 SHSEAERRRRERINNHLAKLRSLLPSTTKTDKASLLAEVIQHVKELK 207
+H EAER+RRE++N LR+++P+ +K DKASLL + I ++ ELK
Sbjct: 188 NHVEAERQRREKLNQRFYALRAVVPNVSKMDKASLLGDAISYITELK 328
>TC8661 weakly similar to UP|O81348 (O81348) Symbiotic ammonium
transporter, partial (34%)
Length = 1007
Score = 52.0 bits (123), Expect = 1e-07
Identities = 38/179 (21%), Positives = 82/179 (45%), Gaps = 21/179 (11%)
Frame = +1
Query: 160 KSHSEAERRRRERINNHLAKLRSLLPSTTKTDKASLLAEVIQHVKELKRQTSLIAETSP- 218
+ H AERRRRE+++ L L +L+P K DKAS+L + I++VK LK + L+ E +
Sbjct: 25 QDHIIAERRRREKLSQSLIALAALIPGLKKMDKASVLGDAIKYVKVLKERLRLLEEQNKN 204
Query: 219 -------------------VPTECDELTVDAAADDEDY-GSNGNKFIIKASLCCDDRSDL 258
+ CD+ T+ + + + + ++ + L C + L
Sbjct: 205 RAMESVVVVNKPQISNDDNSSSSCDDGTIIGSEEALPHVEARVSEKDVLLRLHCKKQKGL 384
Query: 259 LPELIKTLKALRLRTLKADITTLGGRVKNVLFITGEEDDHEYCISSIQEALKAVMEKSV 317
L +++ ++ L L + + + G + ++ + ++ I+ + + L+ +S+
Sbjct: 385 LLKILFEIQNLHLFVVNSSVLPFGDSILDITIVAQMGAEYNLTINELVKNLRVAALRSM 561
>TC9193 similar to GB|AAN28890.1|23506061|AY143951 At4g02590/T10P11_13
{Arabidopsis thaliana;}, partial (59%)
Length = 1474
Score = 50.4 bits (119), Expect = 4e-07
Identities = 25/64 (39%), Positives = 41/64 (64%)
Frame = +1
Query: 151 MEAKALAASKSHSEAERRRRERINNHLAKLRSLLPSTTKTDKASLLAEVIQHVKELKRQT 210
+ A+ A+ HS AER RRERI + + L+PS KTD+A++L E++ +VK L+ Q
Sbjct: 433 VRARRGQATDPHSIAERLRRERIAERIRA*QELVPSVNKTDRAAMLDEIVDYVKFLRLQV 612
Query: 211 SLIA 214
+++
Sbjct: 613 KVLS 624
>BP036451
Length = 511
Score = 49.7 bits (117), Expect = 7e-07
Identities = 35/119 (29%), Positives = 54/119 (44%), Gaps = 1/119 (0%)
Frame = +2
Query: 91 QASPYASLFNRRVPSLQFAYDHHAGSEHLRIISDTLQHGSGSGPFGGFQGEVGKMSAQEI 150
Q P + F SL Y S++ I G G G Q ++G + I
Sbjct: 116 QVPPPPTAFRELFQSLPRGYSFPMSSQNGSIFGGEGDEIDGDG---GSQLDMGVLEFNTI 286
Query: 151 MEAKA-LAASKSHSEAERRRRERINNHLAKLRSLLPSTTKTDKASLLAEVIQHVKELKR 208
A + + E+ RRE++N LRSL+PS TK D+AS++ + I +++EL R
Sbjct: 287 NRVTASVGKGREGKGTEKERREQLNGKYKILRSLIPSPTKIDRASVVGDAIDYIRELIR 463
>CB827320
Length = 522
Score = 48.9 bits (115), Expect = 1e-06
Identities = 21/62 (33%), Positives = 41/62 (65%)
Frame = +1
Query: 152 EAKALAASKSHSEAERRRRERINNHLAKLRSLLPSTTKTDKASLLAEVIQHVKELKRQTS 211
+ A +A+ + +ER RR+++N L LRS++P+ +K DKAS++ + I++++ L Q
Sbjct: 202 DGAASSAASKNIVSERNRRKKLNERLFALRSVVPNISKMDKASIIKDAIEYIEHLHEQEK 381
Query: 212 LI 213
+I
Sbjct: 382 II 387
>CB828026
Length = 542
Score = 47.8 bits (112), Expect = 3e-06
Identities = 26/65 (40%), Positives = 42/65 (64%), Gaps = 1/65 (1%)
Frame = +2
Query: 151 MEAKALAASKSHSEAERRRRERINNHLAKLRSLLPSTTK-TDKASLLAEVIQHVKELKRQ 209
+ A+ A+ SHS AER RRE+I+ + L+ L+P +K T KA +L E+I +V+ L+RQ
Sbjct: 116 VRARRGQATNSHSLAERVRREKISERMKFLQDLVPGCSKVTGKAVMLDEIINYVQSLQRQ 295
Query: 210 TSLIA 214
++
Sbjct: 296 VEFLS 310
>TC17437
Length = 455
Score = 47.8 bits (112), Expect = 3e-06
Identities = 31/80 (38%), Positives = 45/80 (55%), Gaps = 7/80 (8%)
Frame = +1
Query: 266 LKALRLRTLKADITTLGGRVKNVLFITGEE----DDHEY---CISSIQEALKAVMEKSVG 318
L +L L +KADI TLG R+KNVL I E +D EY S+ AL++V+ +
Sbjct: 19 LDSLHLMIIKADIATLGSRMKNVLVIISCEERNFEDAEYRQFLAGSVHPALRSVLHRFCV 198
Query: 319 DESASGSVKRQRTNIISISN 338
+ G+ KR+R +I S S+
Sbjct: 199 SQEVIGTQKRRRISIFSSSS 258
>TC18748 similar to GB|AAN28890.1|23506061|AY143951 At4g02590/T10P11_13
{Arabidopsis thaliana;}, partial (36%)
Length = 803
Score = 40.8 bits (94), Expect = 3e-04
Identities = 21/46 (45%), Positives = 29/46 (62%)
Frame = +1
Query: 151 MEAKALAASKSHSEAERRRRERINNHLAKLRSLLPSTTKTDKASLL 196
+ A+ A+ HS AER RRERI + L+ L+PS KTD+A +L
Sbjct: 463 VRARRGQATDPHSIAERLRRERIAERIRALQELVPSVNKTDRAVML 600
>BI419711
Length = 539
Score = 38.1 bits (87), Expect = 0.002
Identities = 19/46 (41%), Positives = 31/46 (67%), Gaps = 2/46 (4%)
Frame = +1
Query: 161 SHSEAERRRRERINNHLAKLRSLLPST--TKTDKASLLAEVIQHVK 204
+H ER RR+++N +L+ LRSL+PS+ + D+AS++ I VK
Sbjct: 400 THIAVERNRRKQMNEYLSALRSLMPSSYVQRGDQASIIGGAINFVK 537
>BP066258
Length = 475
Score = 36.2 bits (82), Expect = 0.008
Identities = 31/119 (26%), Positives = 50/119 (41%), Gaps = 1/119 (0%)
Frame = -1
Query: 93 SPYASLFN-RRVPSLQFAYDHHAGSEHLRIISDTLQHGSGSGPFGGFQGEVGKMSAQEIM 151
+P ASL N V S ++ DH + ++ + H S G E + S++
Sbjct: 427 NPEASLHNVHSVNSKAYSIDHPS------LLREDKHHSLSSSTCPGSYSEQFERSSEPSK 266
Query: 152 EAKALAASKSHSEAERRRRERINNHLAKLRSLLPSTTKTDKASLLAEVIQHVKELKRQT 210
+K A R R+ I + + LR L+P+ K SLL I+H+ L+ T
Sbjct: 265 NSKKRARPGESCRPRPRDRQLIQDRIKDLRELVPNGAKCSIDSLLERAIKHMLFLQSVT 89
>BI420376
Length = 562
Score = 35.8 bits (81), Expect = 0.011
Identities = 25/74 (33%), Positives = 30/74 (39%), Gaps = 8/74 (10%)
Frame = +3
Query: 69 MQQLHHHH-HPFNPHDPFVIP--QQQASPYASLFNRRVPSLQFAYDHHAGSEHLRIISDT 125
+ LHHH HP PH P P + L R++P QF HH H R +
Sbjct: 102 LHYLHHHSLHPLLPHRPLATPNKHRHRRRRQPLLRRQLPPPQFHLHHHF-LRHPRFLRPP 278
Query: 126 LQ-----HGSGSGP 134
LQ HG G P
Sbjct: 279 LQPRPAAHGGGCHP 320
Score = 26.2 bits (56), Expect = 8.3
Identities = 14/36 (38%), Positives = 18/36 (49%)
Frame = +3
Query: 61 PILQQTWSMQQLHHHHHPFNPHDPFVIPQQQASPYA 96
P+L++ Q H HHH F H F+ P Q P A
Sbjct: 195 PLLRRQLPPPQFHLHHH-FLRHPRFLRPPLQPRPAA 299
>TC10830 weakly similar to UP|Q94JL9 (Q94JL9) At1g68400/T2E12_5, partial
(24%)
Length = 702
Score = 34.7 bits (78), Expect = 0.023
Identities = 21/62 (33%), Positives = 30/62 (47%)
Frame = +3
Query: 69 MQQLHHHHHPFNPHDPFVIPQQQASPYASLFNRRVPSLQFAYDHHAGSEHLRIISDTLQH 128
+QQ HHHHH F+ H F +SP +SL +P Q + H + S+ L H
Sbjct: 96 LQQCHHHHHLFSHHSSF--SHSPSSPSSSLHKPPLPPTQIS-TH*SPSKPLPTPPTNSPH 266
Query: 129 GS 130
G+
Sbjct: 267 GT 272
>AV772495
Length = 572
Score = 32.7 bits (73), Expect = 0.089
Identities = 16/32 (50%), Positives = 21/32 (65%)
Frame = -3
Query: 158 ASKSHSEAERRRRERINNHLAKLRSLLPSTTK 189
A+ SHS AER RRE+IN + L+ L+P K
Sbjct: 435 ATDSHSLAERARREKINARMKLLQELVPGCYK 340
>TC18477
Length = 655
Score = 32.0 bits (71), Expect = 0.15
Identities = 18/58 (31%), Positives = 28/58 (48%)
Frame = +3
Query: 162 HSEAERRRRERINNHLAKLRSLLPSTTKTDKASLLAEVIQHVKELKRQTSLIAETSPV 219
H E ER+RR+ + + A LRSLLP K SL + + +K + + E +
Sbjct: 294 HREIERKRRQDMASFYASLRSLLPLEFIKGKRSLSEHINEAANYIKHMQNNLKELGAI 467
>TC9210
Length = 944
Score = 32.0 bits (71), Expect = 0.15
Identities = 15/47 (31%), Positives = 22/47 (45%)
Frame = +2
Query: 72 LHHHHHPFNPHDPFVIPQQQASPYASLFNRRVPSLQFAYDHHAGSEH 118
+HHHHHPF F + +P+ S N +L Y+ A + H
Sbjct: 113 IHHHHHPFQLRFFFNLNDLPQNPFPSFQNHTTTNLYQ*YNFTAHNTH 253
>BP038129
Length = 558
Score = 30.4 bits (67), Expect = 0.44
Identities = 20/72 (27%), Positives = 31/72 (42%), Gaps = 1/72 (1%)
Frame = +2
Query: 180 LRSLLPSTTKTDKASLLAEVIQHVKELKRQTSLIAETSPVP-TECDELTVDAAADDEDYG 238
+R ++ +K S +QH+ Q +L A+T P+P D +TV A G
Sbjct: 140 IRCASSNSDSGNKVSSRLSQVQHLLHEAEQRALSADTGPIPKITLDHVTVSFARSGGPGG 319
Query: 239 SNGNKFIIKASL 250
N NK K +
Sbjct: 320 QNVNKVNTKVDM 355
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.315 0.130 0.370
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,808,278
Number of Sequences: 28460
Number of extensions: 90781
Number of successful extensions: 1601
Number of sequences better than 10.0: 114
Number of HSP's better than 10.0 without gapping: 1332
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1512
length of query: 338
length of database: 4,897,600
effective HSP length: 91
effective length of query: 247
effective length of database: 2,307,740
effective search space: 570011780
effective search space used: 570011780
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 55 (25.8 bits)
Medicago: description of AC146570.4


