

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146564.2 - phase: 0
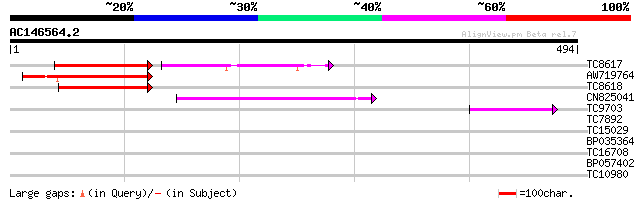
(494 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC8617 similar to UP|Q9SSZ4 (Q9SSZ4) Asparaginyl endopeptidase, ... 123 4e-38
AW719764 138 2e-33
TC8618 weakly similar to UP|Q852T2 (Q852T2) Vacuolar processing ... 113 8e-26
CN825041 89 1e-18
TC9703 similar to UP|Q9XFZ4 (Q9XFZ4) Asparaginyl endopeptidase (... 41 4e-04
TC7892 weakly similar to UP|Q9SSZ4 (Q9SSZ4) Asparaginyl endopept... 36 0.016
TC15029 weakly similar to UP|Q9XFZ4 (Q9XFZ4) Asparaginyl endopep... 33 0.081
BP035364 31 0.53
TC16708 30 1.2
BP057402 27 5.8
TC10980 similar to UP|Q8VZA1 (Q8VZA1) Laccase (Diphenol oxidase)... 27 10.0
>TC8617 similar to UP|Q9SSZ4 (Q9SSZ4) Asparaginyl endopeptidase, partial
(44%)
Length = 904
Score = 123 bits (309), Expect(2) = 4e-38
Identities = 58/85 (68%), Positives = 65/85 (76%)
Frame = +3
Query: 40 VVDDAEVDEVGTRWAVLVAGSSGYGNYRHQADVCHAYQLLIKGGVKEENIVVFMYDDIAN 99
V D + G RWA+LVAGS GYGNYRHQADVCHAYQ+L KGG+ +ENI+VFMYDDIAN
Sbjct: 165 VTGDHQSSTEGKRWALLVAGSYGYGNYRHQADVCHAYQILKKGGLPDENIIVFMYDDIAN 344
Query: 100 NELNPRPGVIINHPQGPNVYVGVPK 124
N+ N PG I N GPNVY GVPK
Sbjct: 345 NKENKWPGKIFNKRNGPNVYEGVPK 419
Score = 51.6 bits (122), Expect(2) = 4e-38
Identities = 52/158 (32%), Positives = 74/158 (45%), Gaps = 8/158 (5%)
Frame = +2
Query: 133 LHWRQRDS*EPIRCYSWRQE*SKGWKWQGHQQQIRR*DIHILLRPWRPWSSWDAK---YA 189
L+WR+ + + +RC W+Q+ W+G Q RR H+L P + + A+ Y+
Sbjct: 422 LYWRRGECKKLLRCA*WQQKCYHRR*WEGFGQWSRRHRFHLLHGPRQYRHNRHAR*YDYS 601
Query: 190 VCICYGFYRRFEEETCIWGLQKDGCIH*SL*KW*HV*GCHAKGS*CLCDNCIQCTRE*LG 249
C R EEETC + KDG I S+* W HV* ++ LC++C +C *LG
Sbjct: 602 RRAC----RCVEEETCRKFI*KDGDILGSV*IWEHV*RTSSR*HKHLCNHCCKCRGR*LG 769
Query: 250 -----NILSWGGTCPTSRVHHLPWGFV*CCLDGRQ*VA 282
+ W G P RV +DGRQ* A
Sbjct: 770 M*LL*RL*RW*GR-PLQRV-----------MDGRQ*QA 847
>AW719764
Length = 414
Score = 138 bits (348), Expect = 2e-33
Identities = 74/117 (63%), Positives = 86/117 (73%), Gaps = 4/117 (3%)
Frame = +3
Query: 12 LLLLLTLDGLVARPNHLEWDPVIRLPGEV---VDDAEVDE-VGTRWAVLVAGSSGYGNYR 67
L LL + +VA L D V+RLP + + + DE GTRWA+L+AGS+GY NYR
Sbjct: 27 LTLLFLIATVVAGGRDLPGD-VLRLPSQASRFLKPSSADENEGTRWAILIAGSNGYWNYR 203
Query: 68 HQADVCHAYQLLIKGGVKEENIVVFMYDDIANNELNPRPGVIINHPQGPNVYVGVPK 124
HQADVCHAYQLL KGG+KEENI+VFMYDDIA NE NPRPGVIIN P G +VY GVPK
Sbjct: 204 HQADVCHAYQLLRKGGLKEENIIVFMYDDIAFNEENPRPGVIINSPHGNDVYEGVPK 374
>TC8618 weakly similar to UP|Q852T2 (Q852T2) Vacuolar processing enzyme-1b,
partial (25%)
Length = 503
Score = 113 bits (282), Expect = 8e-26
Identities = 53/82 (64%), Positives = 61/82 (73%)
Frame = +2
Query: 43 DAEVDEVGTRWAVLVAGSSGYGNYRHQADVCHAYQLLIKGGVKEENIVVFMYDDIANNEL 102
D + G RWA LVAGS+ + NYRHQAD+CHAYQ+L KGG+ ENI+VFMYDDIA N+
Sbjct: 140 DHQSSTEGKRWAWLVAGSNEFYNYRHQADICHAYQILKKGGLPNENIIVFMYDDIAYNKE 319
Query: 103 NPRPGVIINHPQGPNVYVGVPK 124
N PG I N P GPNVY GVPK
Sbjct: 320 NKWPGKIFNSPNGPNVYEGVPK 385
>CN825041
Length = 673
Score = 89.4 bits (220), Expect = 1e-18
Identities = 59/174 (33%), Positives = 88/174 (49%)
Frame = +3
Query: 146 CYSWRQE*SKGWKWQGHQQQIRR*DIHILLRPWRPWSSWDAKYAVCICYGFYRRFEEETC 205
CY+W+ W+W+G I+IL R W W+A ++ +C EEE C
Sbjct: 21 CYTWK*VGCYRWQWEGC**WS**SYIYILQRSWGSRGPWNAH*SLLVCI*SD*SLEEEAC 200
Query: 206 IWGLQKDGCIH*SL*KW*HV*GCHAKGS*CLCDNCIQCTRE*LGNILSWGGTCPTSRVHH 265
W + K + L* W ++* + LC N +C R+ LGNILSW + ++R+ +
Sbjct: 201 FWNI*KPSILSRGL*IWEYL*RSSS*RFEYLCYNSFKCGRKQLGNILSWRES*SSARI*N 380
Query: 266 LPWGFV*CCLDGRQ*VAQSKKGNGETTIQVG*RTDFKLQQLCIRLSCDAIW*YQ 319
LP V CC+DGRQ* Q N TI +G D++ + + CDAIW*++
Sbjct: 381 LPG*PVQCCVDGRQ*YTQFAIRNSAPTIPIGETKDYE-WKYNLWFPCDAIW*HK 539
>TC9703 similar to UP|Q9XFZ4 (Q9XFZ4) Asparaginyl endopeptidase (VmPE-1),
partial (28%)
Length = 727
Score = 41.2 bits (95), Expect = 4e-04
Identities = 27/77 (35%), Positives = 38/77 (49%)
Frame = +2
Query: 401 WSFTVWSNKRFFSSTGRKSDWPTSC**LGMLKIKGSVV*NSLWVTDPVWDETHASICKHL 460
W +W K S+ + T+C**LG+ G *+ LW+ VWDETH +C+ L
Sbjct: 128 WEALIWHGKGSRSAQQC*TCRATTC**LGLP*NSG*DF*DILWIPFSVWDETHEVLCQLL 307
Query: 461 QQWYFRGFYGEGMHGSM 477
Q+ G G+ SM
Sbjct: 308 QRRNTERANG*GLGTSM 358
>TC7892 weakly similar to UP|Q9SSZ4 (Q9SSZ4) Asparaginyl endopeptidase,
partial (17%)
Length = 677
Score = 35.8 bits (81), Expect = 0.016
Identities = 20/61 (32%), Positives = 28/61 (45%)
Frame = +1
Query: 405 VWSNKRFFSSTGRKSDWPTSC**LGMLKIKGSVV*NSLWVTDPVWDETHASICKHLQQWY 464
VW ++ S SC**LGM +* +L +W E H +C+H+Q W
Sbjct: 196 VWRRQKLQHHGSCSSGRAASC**LGMF*DADENL*EALRRLIKLWKEVHKGLCQHVQCWN 375
Query: 465 F 465
F
Sbjct: 376 F 378
>TC15029 weakly similar to UP|Q9XFZ4 (Q9XFZ4) Asparaginyl endopeptidase
(VmPE-1), partial (10%)
Length = 624
Score = 33.5 bits (75), Expect = 0.081
Identities = 20/61 (32%), Positives = 26/61 (41%)
Frame = +2
Query: 405 VWSNKRFFSSTGRKSDWPTSC**LGMLKIKGSVV*NSLWVTDPVWDETHASICKHLQQWY 464
VW ++ S SC**LGM + +L VW E H +C H+Q W
Sbjct: 185 VWRRQKLQHHGSCSSGRAASC**LGMF*DTDKNLQEALRSFIKVWKEIHKGLCLHVQCWN 364
Query: 465 F 465
F
Sbjct: 365 F 367
>BP035364
Length = 556
Score = 30.8 bits (68), Expect = 0.53
Identities = 22/82 (26%), Positives = 31/82 (36%), Gaps = 4/82 (4%)
Frame = +3
Query: 59 GSSGYGNYRHQADVCHAYQLLIKGGVKEENIVVFMYDDIANNEL----NPRPGVIINHPQ 114
GS +GN+R CH + I G F + + L NP+ V ++ P
Sbjct: 207 GSVDHGNHRDDGSQCHDHSSEIHSGENRHGYSFFSFSHGDSPSLSGLKNPQISVQLSPPL 386
Query: 115 GPNVYVGVPKIFFFNKPRLHWR 136
PN + F F R WR
Sbjct: 387 LPNQH------FSFRSNRFQWR 434
>TC16708
Length = 515
Score = 29.6 bits (65), Expect = 1.2
Identities = 15/35 (42%), Positives = 22/35 (62%)
Frame = +3
Query: 30 WDPVIRLPGEVVDDAEVDEVGTRWAVLVAGSSGYG 64
W V R+PGE+ + A V + +VL+ GS+GYG
Sbjct: 186 WQKVARVPGEICNVAYVGALD--GSVLLIGSNGYG 284
>BP057402
Length = 400
Score = 27.3 bits (59), Expect = 5.8
Identities = 11/25 (44%), Positives = 17/25 (68%)
Frame = -3
Query: 187 KYAVCICYGFYRRFEEETCIWGLQK 211
K++ C C G R F E++C+ GLQ+
Sbjct: 341 KWSNCPCCGTIRFFPEKSCLEGLQQ 267
>TC10980 similar to UP|Q8VZA1 (Q8VZA1) Laccase (Diphenol oxidase)-like
protein, partial (22%)
Length = 572
Score = 26.6 bits (57), Expect = 10.0
Identities = 8/19 (42%), Positives = 12/19 (63%)
Frame = +1
Query: 179 RPWSSWDAKYAVCICYGFY 197
R W W+ +Y+ C CY +Y
Sbjct: 385 RAWIVWNNQYSQCNCYFYY 441
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.348 0.155 0.614
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,714,108
Number of Sequences: 28460
Number of extensions: 204800
Number of successful extensions: 1795
Number of sequences better than 10.0: 23
Number of HSP's better than 10.0 without gapping: 1766
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1791
length of query: 494
length of database: 4,897,600
effective HSP length: 94
effective length of query: 400
effective length of database: 2,222,360
effective search space: 888944000
effective search space used: 888944000
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 14 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.8 bits)
S2: 57 (26.6 bits)
Medicago: description of AC146564.2


