

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146561.5 + phase: 2 /pseudo/partial
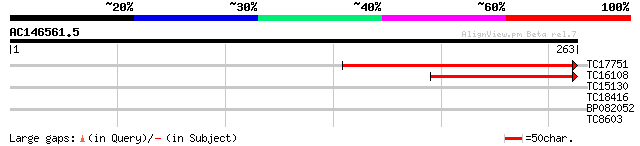
(263 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC17751 weakly similar to UP|Q7XHN9 (Q7XHN9) Tetratricopeptide r... 167 2e-42
TC16108 weakly similar to UP|Q7XHN9 (Q7XHN9) Tetratricopeptide r... 124 2e-29
TC15130 similar to UP|O81488 (O81488) F9D12.13 protein, partial ... 28 2.1
TC18416 26 6.1
BP082052 26 8.0
TC8603 similar to UP|Q8LBJ5 (Q8LBJ5) Nucleic acid binding protei... 26 8.0
>TC17751 weakly similar to UP|Q7XHN9 (Q7XHN9) Tetratricopeptide
repeat(TPR)-containing protein-like, partial (8%)
Length = 550
Score = 167 bits (423), Expect = 2e-42
Identities = 83/109 (76%), Positives = 93/109 (85%)
Frame = +3
Query: 155 LENVRDAILRMTYYWYNFMPLSRGTAAVGFAVMLGLLLAANMEFTGSIPQGFQVDWEAIL 214
LE+VR+AILRMTYYWYNF PL +G+AAVGF VM GLLLAA+MEFTGSIPQG QVD EAIL
Sbjct: 3 LEDVRNAILRMTYYWYNFRPLYKGSAAVGFVVMQGLLLAAHMEFTGSIPQGLQVDLEAIL 182
Query: 215 NLDPNSFVDSVKSWLYPSLKVTTSWKDYHDVASTFATTGSVVAALSSYD 263
+ D NSFVDSV++WLYPSLK+TT WK Y D+ STF T GSVVAALS D
Sbjct: 183 SSDTNSFVDSVETWLYPSLKLTTLWKGYPDIVSTFETIGSVVAALSFSD 329
>TC16108 weakly similar to UP|Q7XHN9 (Q7XHN9) Tetratricopeptide
repeat(TPR)-containing protein-like, partial (3%)
Length = 597
Score = 124 bits (310), Expect = 2e-29
Identities = 58/68 (85%), Positives = 64/68 (93%)
Frame = +3
Query: 196 MEFTGSIPQGFQVDWEAILNLDPNSFVDSVKSWLYPSLKVTTSWKDYHDVASTFATTGSV 255
MEFTGSIPQGFQVDWEAILN+D NSF++SV+SWLYPSLKVTTSWKDY D+ASTF TTGSV
Sbjct: 3 MEFTGSIPQGFQVDWEAILNMDLNSFMNSVQSWLYPSLKVTTSWKDYPDIASTFTTTGSV 182
Query: 256 VAALSSYD 263
+AALSS D
Sbjct: 183 IAALSSSD 206
>TC15130 similar to UP|O81488 (O81488) F9D12.13 protein, partial (26%)
Length = 583
Score = 27.7 bits (60), Expect = 2.1
Identities = 16/37 (43%), Positives = 20/37 (53%), Gaps = 4/37 (10%)
Frame = +3
Query: 125 EDYDAEMAMAWEALCNAYCGENYGSTDF----DVLEN 157
ED + + E LC A CGENY S +F D+ EN
Sbjct: 93 EDEEEDDEEHGETLCGA-CGENYASDEFWICCDICEN 200
>TC18416
Length = 426
Score = 26.2 bits (56), Expect = 6.1
Identities = 14/47 (29%), Positives = 24/47 (50%)
Frame = -3
Query: 140 NAYCGENYGSTDFDVLENVRDAILRMTYYWYNFMPLSRGTAAVGFAV 186
N + GE G+ D D +E + + + W++F L R + GF+V
Sbjct: 157 NVFSGET*GAED-DEVEAIAAVGIGIHVVWFHFWRLQRRRVSSGFSV 20
>BP082052
Length = 371
Score = 25.8 bits (55), Expect = 8.0
Identities = 8/17 (47%), Positives = 13/17 (76%)
Frame = -1
Query: 234 KVTTSWKDYHDVASTFA 250
K+ TSW ++HDV S ++
Sbjct: 131 KMRTSWTEFHDVTSNYS 81
>TC8603 similar to UP|Q8LBJ5 (Q8LBJ5) Nucleic acid binding protein-like,
partial (88%)
Length = 1232
Score = 25.8 bits (55), Expect = 8.0
Identities = 10/17 (58%), Positives = 14/17 (81%)
Frame = +1
Query: 136 EALCNAYCGENYGSTDF 152
+ALC A CG+NYG+ +F
Sbjct: 718 DALCGA-CGDNYGTDEF 765
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.319 0.134 0.419
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,954,636
Number of Sequences: 28460
Number of extensions: 66900
Number of successful extensions: 312
Number of sequences better than 10.0: 12
Number of HSP's better than 10.0 without gapping: 312
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 312
length of query: 263
length of database: 4,897,600
effective HSP length: 88
effective length of query: 175
effective length of database: 2,393,120
effective search space: 418796000
effective search space used: 418796000
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 54 (25.4 bits)
Medicago: description of AC146561.5


