

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146560.10 + phase: 0
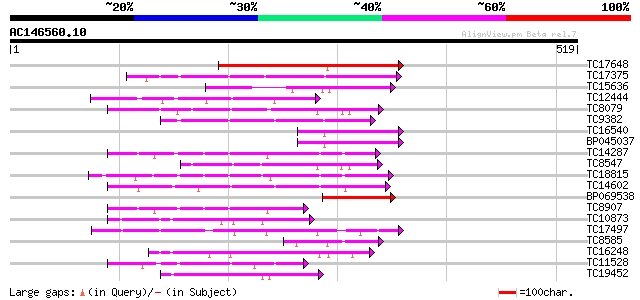
(519 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC17648 similar to UP|Q9AYN9 (Q9AYN9) NQK1 MAPKK, partial (51%) 174 2e-44
TC17375 mitogen-activated kinase kinase kinase alpha [Lotus corn... 142 1e-34
TC15636 similar to UP|Q93WR7 (Q93WR7) MAP kinase kinase, partial... 141 2e-34
TC12444 similar to UP|O22981 (O22981) Ser/Thr protein kinase iso... 93 1e-19
TC8079 homologue to UP|Q9M6S1 (Q9M6S1) MAP kinase 3, complete 92 2e-19
TC9382 weakly similar to UP|ABL_DROME (P00522) Tyrosine-protein ... 91 6e-19
TC16540 similar to UP|Q93WR7 (Q93WR7) MAP kinase kinase, partial... 89 2e-18
BP045037 89 2e-18
TC14287 similar to UP|Q9LEU7 (Q9LEU7) Serine/threonine protein k... 86 2e-17
TC8547 homologue to UP|MMK1_MEDSA (Q07176) Mitogen-activated pro... 86 2e-17
TC18815 similar to UP|Q8W1D5 (Q8W1D5) CBL-interacting protein ki... 78 4e-15
TC14602 UP|Q8GS56 (Q8GS56) Phosphoenolpyruvate carboxylase kinas... 77 7e-15
BP069538 74 6e-14
TC8907 similar to UP|Q8H0X3 (Q8H0X3) Serine/threonine protein ki... 74 8e-14
TC10873 similar to UP|Q9LDZ5 (Q9LDZ5) F2D10.13 (F5M15.3), partia... 70 6e-13
TC17497 UP|CAE45595 (CAE45595) S-receptor kinase-like protein 2,... 70 8e-13
TC8585 weakly similar to PIR|T51339|T51339 mitogen-activated pro... 70 8e-13
TC16248 similar to UP|BAC87845 (BAC87845) Leucine-rich repeat re... 69 2e-12
TC11528 homologue to UP|Q39868 (Q39868) Protein kinase , partial... 69 2e-12
TC19452 homologue to UP|Q8S9J9 (Q8S9J9) At1g14000/F7A19_9, parti... 68 3e-12
>TC17648 similar to UP|Q9AYN9 (Q9AYN9) NQK1 MAPKK, partial (51%)
Length = 741
Score = 174 bits (442), Expect = 2e-44
Identities = 88/175 (50%), Positives = 115/175 (65%), Gaps = 6/175 (3%)
Frame = +3
Query: 192 LRGLSYLHGVRYLVHRDIKPANLLVNLKGEPKITDFGISAGLENSVAMCATFVGTVTYMS 251
L+GL YLH R+++HRD KP++LLVN +GE KITDFG+SA L S+ TFVGT YMS
Sbjct: 3 LQGLVYLHNERHVIHRDSKPSHLLVNHQGEVKITDFGVSAMLATSMGQRDTFVGTYNYMS 182
Query: 252 PERIRNESYSYPADIWSLGLALLESGTGEFPYTANEGP------VNLMLQILDDPSPSPS 305
P RI +Y Y +DIWSLG+ +LE G FPY +E L+ I++ P PS
Sbjct: 183 PARISGSTYDYSSDIWSLGMVVLECAIGRFPYIQSEDQQAWPSFYELLAAIVESPPPSAP 362
Query: 306 KEKFSPEFCSFVDACLQKDPDNRPTAEQLLLHPFITKYETAKVDLPGFVRSVFDP 360
++FSPEFCSFV +C+QKDP +R T+ +LL HPFI K+E +DL V S+ P
Sbjct: 363 SDQFSPEFCSFVSSCIQKDPQDRLTSLELLDHPFIKKFEDKDLDLEILVGSLEPP 527
>TC17375 mitogen-activated kinase kinase kinase alpha [Lotus corniculatus
var. japonicus]
Length = 1422
Score = 142 bits (359), Expect = 1e-34
Identities = 91/257 (35%), Positives = 145/257 (56%), Gaps = 6/257 (2%)
Frame = +3
Query: 108 RVIALKKINIFEKEKR-----QQLLTEIRTLCEAPCYEGLVEFHGAFYTPDSGQISIALE 162
++ A+K++ +F +K +QL EI L + + +V+++G+ +S +S+ LE
Sbjct: 3 QMCAIKEVKVFSDDKTSKECLKQLNQEINLLNQFS-HPNIVQYYGSELGEES--LSVYLE 173
Query: 163 YMDGGSLADILRMHRTIPEPILSSMFQKLLRGLSYLHGVRYLVHRDIKPANLLVNLKGEP 222
Y+ GGS+ +L+ + EP++ + ++++ GL+YLH R VHRDIK AN+LV+ GE
Sbjct: 174 YVSGGSIHKLLQEYGAFKEPVIQNYTRQIVSGLAYLHS-RNTVHRDIKGANILVDPNGEI 350
Query: 223 KITDFGISAGLENSVAMCATFVGTVTYMSPERIRNES-YSYPADIWSLGLALLESGTGEF 281
K+ DFG+S + NS A +F G+ +M+PE + N + Y P DI SLG +LE T +
Sbjct: 351 KLADFGMSKHI-NSAASMLSFKGSPYWMAPEVVMNTNGYGLPVDISSLGCTILEMATSKP 527
Query: 282 PYTANEGPVNLMLQILDDPSPSPSKEKFSPEFCSFVDACLQKDPDNRPTAEQLLLHPFIT 341
P++ EG V + +I + E S + +F+ CLQ+DP RPTA+ LL HPFI
Sbjct: 528 PWSQFEG-VAAIFKIGNSKDMPEIPEHLSDDAKNFIKQCLQRDPLARPTAQSLLNHPFIR 704
Query: 342 KYETAKVDLPGFVRSVF 358
KV R F
Sbjct: 705 DQSATKVANASITRDAF 755
>TC15636 similar to UP|Q93WR7 (Q93WR7) MAP kinase kinase, partial (46%)
Length = 1184
Score = 141 bits (356), Expect = 2e-34
Identities = 77/182 (42%), Positives = 109/182 (59%), Gaps = 8/182 (4%)
Frame = +2
Query: 180 PEPILSSMFQKLLRGLSYLHGVRYLVHRDIKPANLLVNLKGEPKITDFGISAGLENSVAM 239
PEP L+S+ +++L+GL YLH ++++HRD+KP+NLL+N +GEP
Sbjct: 2 PEPYLASICKQVLKGLMYLHHEKHIIHRDLKPSNLLINHRGEP----------------- 130
Query: 240 CATFVGTVTYMSPERIRN--ESYSYPADIWSLGLALLESGTGEFPYTA---NEGPVN--- 291
ERI + Y+Y +DIWSLGL L TG FPYT +EG N
Sbjct: 131 -------------ERINGSQDGYNYKSDIWSLGLMFLRCATGMFPYTPPDQSEGWENIYQ 271
Query: 292 LMLQILDDPSPSPSKEKFSPEFCSFVDACLQKDPDNRPTAEQLLLHPFITKYETAKVDLP 351
L+ I+++PSPS S ++FS EFCSF+ ACLQK+P +RP+A +L+ HPFI Y+ VDL
Sbjct: 272 LIEAIVENPSPSVSSDEFSSEFCSFIAACLQKNPKDRPSAPELMRHPFINMYDDLAVDLS 451
Query: 352 GF 353
+
Sbjct: 452 AY 457
>TC12444 similar to UP|O22981 (O22981) Ser/Thr protein kinase isolog,
partial (37%)
Length = 709
Score = 92.8 bits (229), Expect = 1e-19
Identities = 60/219 (27%), Positives = 114/219 (51%), Gaps = 9/219 (4%)
Frame = +2
Query: 75 KTYRCGSHEMRIFGAIGSGASSVVQRAMHIPTHRVIALKKINIFEKEKRQQLLTEIRTLC 134
+TY S + + +G GAS+ V RA+ +P +A+K +++ ++ L +IR
Sbjct: 62 RTYSLNSGDYNLLEEVGYGASATVYRAVFLPRDEEVAVKCVDL---DRCNANLEDIRREA 232
Query: 135 EAPC---YEGLVEFHGAFYTPDSGQISIALEYMDGGSLADILRMHRT--IPEPILSSMFQ 189
+ + +V H +F ++ + + +M GS ++++ E + S+ +
Sbjct: 233 QTMSLIDHRNVVRAHWSFVV--DRRLWVVMPFMAQGSCLHLMKVAYPDGFEEEAIGSILK 406
Query: 190 KLLRGLSYLHGVRYLVHRDIKPANLLVNLKGEPKITDFGISAGLENSVAMCA---TFVGT 246
+ L+ L YLH + +HRD+K N+L+ G+ K+ DFG+SA + ++ TFVGT
Sbjct: 407 ETLKALEYLHRHGH-IHRDVKAGNILLGSDGQVKLADFGVSASMFDAGERQRNRNTFVGT 583
Query: 247 VTYMSPERIR-NESYSYPADIWSLGLALLESGTGEFPYT 284
+M+PE ++ Y++ AD+WS G+ LE G P++
Sbjct: 584 PCWMAPEVLQPGTGYNFKADVWSFGITALELAHGHAPFS 700
>TC8079 homologue to UP|Q9M6S1 (Q9M6S1) MAP kinase 3, complete
Length = 1550
Score = 92.0 bits (227), Expect = 2e-19
Identities = 82/288 (28%), Positives = 132/288 (45%), Gaps = 35/288 (12%)
Frame = +3
Query: 90 IGSGASSVVQRAMHIPTHRVIALKKI-NIFEKEK-RQQLLTEIRTLCEAPCYEGLVEFHG 147
IG GA +V ++ T+ ++A+KKI N F+ ++ L EI+ L +E ++
Sbjct: 309 IGRGAYGIVCSLLNTETNELVAVKKIANAFDNHMDAKRTLREIKLLRHLD-HENVIALRD 485
Query: 148 AFYTP---DSGQISIALEYMDGGSLADILRMHRTIPEPILSSMFQKLLRGLSYLHGVRYL 204
P + + I E MD L I+R ++ + E ++LRGL Y+H +
Sbjct: 486 VIPPPLRREFTDVYITTELMDT-DLHQIIRSNQGLSEEHCQYFLYQVLRGLKYIHSAN-I 659
Query: 205 VHRDIKPANLLVNLKGEPKITDFGIS-AGLENSVAMCATFVGTVTYMSPERIRNES-YSY 262
+HRD+KP+NLL+N + KI DFG++ +EN +V T Y +PE + N S Y+
Sbjct: 660 IHRDLKPSNLLLNSNCDLKIIDFGLARPTVEND--FMTEYVVTRWYRAPELLLNSSDYTS 833
Query: 263 PADIWSLGLALLESGTGE--FPYTANEGPVNLMLQILDDPSPS----------------- 303
D+WS+G +E + FP + + L+ ++L P+ +
Sbjct: 834 AIDVWSVGCIFMELMNKKPLFPGKDHVHQLRLLTELLGTPTEADLGLVKNDDVRRYIRQL 1013
Query: 304 ------PSKEKF---SPEFCSFVDACLQKDPDNRPTAEQLLLHPFITK 342
P + F P VD L DP R T EQ L HP++ K
Sbjct: 1014PQYPRQPLTKVFPHVHPMAMDLVDKMLTIDPTKRITVEQALAHPYLEK 1157
>TC9382 weakly similar to UP|ABL_DROME (P00522) Tyrosine-protein kinase Abl
(D-ash) , partial (5%)
Length = 1197
Score = 90.5 bits (223), Expect = 6e-19
Identities = 61/199 (30%), Positives = 103/199 (51%), Gaps = 2/199 (1%)
Frame = +2
Query: 139 YEGLVEFHGAFY-TPDSGQISIALEYMDGGSLADILRMHRTIPE-PILSSMFQKLLRGLS 196
++ +V+F GA TP+ + I E+M GSL D L R + + P L + + +G++
Sbjct: 107 HKNVVQFIGACTRTPN---LCIVTEFMSRGSLYDFLHKQRGVFKLPSLLKVAIDVSKGMN 277
Query: 197 YLHGVRYLVHRDIKPANLLVNLKGEPKITDFGISAGLENSVAMCATFVGTVTYMSPERIR 256
YLH ++HRD+K NLL++ K+ DFG++ + S M A GT +M+PE I
Sbjct: 278 YLHQ-NNIIHRDLKTGNLLMDENELVKVADFGVARVITQSGVMTAE-TGTYRWMAPEVIE 451
Query: 257 NESYSYPADIWSLGLALLESGTGEFPYTANEGPVNLMLQILDDPSPSPSKEKFSPEFCSF 316
++ Y AD++S G+AL E TGE PY+ P+ + ++ + P
Sbjct: 452 HKPYDQKADVFSFGIALWELLTGELPYSYLT-PLQAAVGVVQKGLRPSIPKNTHPRLSEL 628
Query: 317 VDACLQKDPDNRPTAEQLL 335
+ C ++DP RP +++
Sbjct: 629 LQRCWKQDPIERPAFSEII 685
>TC16540 similar to UP|Q93WR7 (Q93WR7) MAP kinase kinase, partial (30%)
Length = 672
Score = 89.0 bits (219), Expect = 2e-18
Identities = 45/103 (43%), Positives = 60/103 (57%), Gaps = 6/103 (5%)
Frame = +1
Query: 264 ADIWSLGLALLESGTGEFPYTAN------EGPVNLMLQILDDPSPSPSKEKFSPEFCSFV 317
+DIWSLGL LLE G FPY E L+ ++D P PS E+FS EFCSF+
Sbjct: 1 SDIWSLGLILLECAMGRFPYAPPDQSETWESIFELIETVVDKPPPSAPSEQFSTEFCSFI 180
Query: 318 DACLQKDPDNRPTAEQLLLHPFITKYETAKVDLPGFVRSVFDP 360
ACLQKDP +R +A++L+ PF + Y+ VDL + + P
Sbjct: 181 SACLQKDPKDRLSAQELMRLPFTSMYDDLDVDLSAYFSNAGSP 309
>BP045037
Length = 565
Score = 89.0 bits (219), Expect = 2e-18
Identities = 45/103 (43%), Positives = 60/103 (57%), Gaps = 6/103 (5%)
Frame = -2
Query: 264 ADIWSLGLALLESGTGEFPYTAN------EGPVNLMLQILDDPSPSPSKEKFSPEFCSFV 317
+DIWSLGL LLE G FPY E L+ ++D P PS E+FS EFCSF+
Sbjct: 564 SDIWSLGLILLECAMGRFPYAPPDQSETWESIFELIETVVDKPPPSAPSEQFSTEFCSFI 385
Query: 318 DACLQKDPDNRPTAEQLLLHPFITKYETAKVDLPGFVRSVFDP 360
ACLQKDP +R +A++L+ PF + Y+ VDL + + P
Sbjct: 384 SACLQKDPKDRLSAQELMRLPFTSMYDDLDVDLSAYFSNAGSP 256
>TC14287 similar to UP|Q9LEU7 (Q9LEU7) Serine/threonine protein kinase-like
protein (CBL-interacting protein kinase 5), partial
(75%)
Length = 1999
Score = 85.5 bits (210), Expect = 2e-17
Identities = 70/256 (27%), Positives = 125/256 (48%), Gaps = 6/256 (2%)
Frame = +3
Query: 90 IGSGASSVVQRAMHIPTHRVIALKKINIFEKEKRQQLLTEIR---TLCEAPCYEGLVEFH 146
+G G + V ++ T+ +A+K I EK K+ +L+ +I+ ++ + +VE
Sbjct: 450 LGQGNFAKVYHGRNLATNENVAIKVIKK-EKLKKDRLVKQIKREVSVMRLVRHPHIVELK 626
Query: 147 GAFYTPDSGQISIALEYMDGGSLADILRMHRTIPEPILSSMFQKLLRGLSYLHGVRYLVH 206
T G+I + +EY+ GG L + + + E FQ+L+ + + H R + H
Sbjct: 627 EVMAT--KGKIFLVMEYVKGGELFTKVNKGK-LNEDDARKYFQQLISAVDFCHS-RGVTH 794
Query: 207 RDIKPANLLVNLKGEPKITDFGISAGLE--NSVAMCATFVGTVTYMSPERIRNESY-SYP 263
RD+KP NLL++ + K++DFG+SA E M T GT Y++PE ++ + Y
Sbjct: 795 RDLKPENLLLDENEDLKVSDFGLSALPEQRRDDGMLVTPCGTPAYVAPEVLKKKGYDGSK 974
Query: 264 ADIWSLGLALLESGTGEFPYTANEGPVNLMLQILDDPSPSPSKEKFSPEFCSFVDACLQK 323
ADIWS G+ L +G P+ E + + + P E SP+ + + L
Sbjct: 975 ADIWSCGVILYALLSGYLPF-QGENVMRIYRKAFKAEYEFP--EWISPQAKNLISNLLVA 1145
Query: 324 DPDNRPTAEQLLLHPF 339
DP+ R + +++ P+
Sbjct: 1146DPEKRYSIPEIISDPW 1193
>TC8547 homologue to UP|MMK1_MEDSA (Q07176) Mitogen-activated protein
kinase homolog MMK1 (MAP kinase MSK7) (MAP kinase ERK1)
, partial (71%)
Length = 1098
Score = 85.5 bits (210), Expect = 2e-17
Identities = 65/213 (30%), Positives = 100/213 (46%), Gaps = 28/213 (13%)
Frame = +1
Query: 157 ISIALEYMDGGSLADILRMHRTIPEPILSSMFQKLLRGLSYLHGVRYLVHRDIKPANLLV 216
+ IA E MD L I+R ++ + E ++LRGL Y+H L HRD+KP+NLL+
Sbjct: 64 VYIAYELMDT-DLHQIIRSNQGLSEEHCQYFLYQILRGLKYIHSANVL-HRDLKPSNLLL 237
Query: 217 NLKGEPKITDFGISAGLENSVAMCATFVGTVTYMSPERIRNES-YSYPADIWSLGLALLE 275
N + KI DFG+ A + + +V T Y +PE + N S Y+ D+WS+G +E
Sbjct: 238 NANCDLKICDFGL-ARVTSETDFMTEYVVTRWYRAPELLLNSSDYTAAIDVWSVGCIFME 414
Query: 276 SGTGE--FPYTANEGPVNLMLQILDDPSP----------------------SPSKEKF-- 309
+ FP + + L+++++ PS +EKF
Sbjct: 415 LMDRKPLFPGRDHVHQLRLLMELIGTPSEDDLGFLNENAKRYIRQLPPYRRQSFQEKFPQ 594
Query: 310 -SPEFCSFVDACLQKDPDNRPTAEQLLLHPFIT 341
PE V+ L DP R T E+ L HP++T
Sbjct: 595 VHPEAIDLVEKMLTFDPRKRITVEEALAHPYLT 693
>TC18815 similar to UP|Q8W1D5 (Q8W1D5) CBL-interacting protein kinase
CIPK25, partial (58%)
Length = 979
Score = 77.8 bits (190), Expect = 4e-15
Identities = 75/286 (26%), Positives = 130/286 (45%), Gaps = 7/286 (2%)
Frame = +2
Query: 73 NEKTYRCGSHEMRIFGAIGSGASSVVQRAMHIPTHRVIALK---KINIFEKEKRQQLLTE 129
+E + G +EM +G G + V A I + +A+K K I ++ Q+ E
Sbjct: 11 DEGGHPIGKYEMG--RVLGKGTLAKVYFAKEITSGEGVAIKVMSKARIKKEGMMDQIKRE 184
Query: 130 IRTLCEAPCYEGLVEFHGAFYTPDSGQISIALEYMDGGSLADILRMHRTIPEPILSSMFQ 189
I ++ + +V T +I +EY+ GG L + + + + + FQ
Sbjct: 185 I-SIMRLVRHPNIVNLKEVMATKT--KIFFIMEYIRGGELFAKVAKGK-LKDDLARRYFQ 352
Query: 190 KLLRGLSYLHGVRYLVHRDIKPANLLVNLKGEPKITDFGISAGLENSV---AMCATFVGT 246
+L+ + Y H R + HRD+KP NLL++ K++DFG+S GL + + T GT
Sbjct: 353 QLISAVDYCHS-RGVSHRDLKPENLLLDENENLKVSDFGLS-GLPEQLRQDGLLHTQCGT 526
Query: 247 VTYMSPERIRNESY-SYPADIWSLGLALLESGTGEFPYTANEGPVNLMLQILDDPSPSPS 305
Y++PE +R + Y + D WS G+ L G P+ +E + + ++L P
Sbjct: 527 PAYVAPEVLRKKGYDGFKTDTWSCGVILYALLAGCLPF-QHENLMTMYNKVLRAEFQFP- 700
Query: 306 KEKFSPEFCSFVDACLQKDPDNRPTAEQLLLHPFITKYETAKVDLP 351
FSPE + L DP+ R T ++ + K +A + +P
Sbjct: 701 -PWFSPESKKLISKILVADPNRRITISSIMRVSWFQKGFSASIPIP 835
>TC14602 UP|Q8GS56 (Q8GS56) Phosphoenolpyruvate carboxylase kinase, complete
Length = 1256
Score = 77.0 bits (188), Expect = 7e-15
Identities = 67/267 (25%), Positives = 119/267 (44%), Gaps = 8/267 (2%)
Frame = +2
Query: 90 IGSGASSVVQRAMHIPTHRVIALKKIN---IFEKEKRQQLLTEIRTLCEAPCYEGLVEFH 146
IG G + R H + A+K I+ + + R L E + + + +++
Sbjct: 86 IGRGRFGTIFRCFHPISTDTFAVKLIDKSLLADSTDRHCLENEPKYMSLLSPHPNILQIF 265
Query: 147 GAFYTPDSGQISIALEYMDGGSLAD--ILRMHRTIPEPILSSMFQKLLRGLSYLHGVRYL 204
F D +S+ +E +L D + +IPE + + ++LL +++ H + +
Sbjct: 266 DVF--EDDDVLSMVIELCQPLTLLDRIVAANGTSIPEVEAAGLMKQLLEAVAHCHRLG-V 436
Query: 205 VHRDIKPANLLVNLKGEPKITDFGISAGLENSVAMCATFVGTVTYMSPERIRNESYSYPA 264
HRD+KP N+L G+ K+ DFG + + M VGT Y++PE + Y
Sbjct: 437 AHRDVKPDNVLFGGGGDLKLADFGSAEWFGDGRRMSGV-VGTPYYVAPEVLMGREYGEKV 613
Query: 265 DIWSLGLALLESGTGEFPYTANEGPVNLMLQILDDPSPSPSK--EKFSPEFCSFVDACLQ 322
D+WS G+ L +G P+ + + ++ PS+ SP + +
Sbjct: 614 DVWSCGVILYIMLSGTPPF-YGDSAAEIFEAVIRGNLRFPSRIFRNVSPAAKDLLRKMIC 790
Query: 323 KDPDNRPTAEQLLLHP-FITKYETAKV 348
+DP NR +AEQ L HP F++ +T V
Sbjct: 791 RDPSNRISAEQALRHPWFLSAGDTVNV 871
>BP069538
Length = 532
Score = 73.9 bits (180), Expect = 6e-14
Identities = 33/67 (49%), Positives = 47/67 (69%)
Frame = -3
Query: 287 EGPVNLMLQILDDPSPSPSKEKFSPEFCSFVDACLQKDPDNRPTAEQLLLHPFITKYETA 346
E L+ I+++PSPS S ++FS EFCSF+ ACLQK+P +RP+A +L+ HPFI Y+
Sbjct: 530 ENIYQLIEAIVENPSPSVSSDEFSSEFCSFIAACLQKNPKDRPSAPELMRHPFINMYDDL 351
Query: 347 KVDLPGF 353
VDL +
Sbjct: 350 AVDLSAY 330
>TC8907 similar to UP|Q8H0X3 (Q8H0X3) Serine/threonine protein kinase-like
protein, partial (44%)
Length = 941
Score = 73.6 bits (179), Expect = 8e-14
Identities = 57/190 (30%), Positives = 100/190 (52%), Gaps = 6/190 (3%)
Frame = +3
Query: 90 IGSGASSVVQRAMHIPTHRVIALKKINIFEKEKRQQLLTEIR---TLCEAPCYEGLVEFH 146
+G G + V ++ T+ +A+K I E+ K+++L+ +I+ ++ + +VE
Sbjct: 360 LGQGNFAKVYHGRNMETNESVAIKVIKK-ERLKKERLVKQIKREVSVMRLVRHPHIVELK 536
Query: 147 GAFYTPDSGQISIALEYMDGGSLADILRMHRTIPEPILSSMFQKLLRGLSYLHGVRYLVH 206
T +I + +EY+ GG L L + + E FQ+L+ + + H R + H
Sbjct: 537 EVMATKT--KIFMVVEYVKGGELFAKLTKGK-MTEVAARKYFQQLISAVDFCHS-RGVTH 704
Query: 207 RDIKPANLLVNLKGEPKITDFGISAGLE--NSVAMCATFVGTVTYMSPERIRNESY-SYP 263
RD+KP NLL++ + K++DFG+S+ E S M T GT Y++PE ++ + Y
Sbjct: 705 RDLKPENLLLDDNEDLKVSDFGLSSLPEQRRSDGMLLTPCGTPAYVAPEVLKKKGYDGSK 884
Query: 264 ADIWSLGLAL 273
ADIWS G+ L
Sbjct: 885 ADIWSCGVIL 914
>TC10873 similar to UP|Q9LDZ5 (Q9LDZ5) F2D10.13 (F5M15.3), partial (77%)
Length = 1478
Score = 70.5 bits (171), Expect = 6e-13
Identities = 59/198 (29%), Positives = 98/198 (48%), Gaps = 8/198 (4%)
Frame = +1
Query: 90 IGSGASSVVQRAMHIPTHRVIALKKINIFEKEKRQQLLTEIRTLCEAPCYEGLVEFHGAF 149
IG G V + + T +A+K+++ ++ Q+ + E+ L + LV G
Sbjct: 469 IGEGGFGKVYKG-RLTTGEAVAVKQLSHDGRQGFQEFVMEVLMLSLLH-HTNLVRLIG-- 636
Query: 150 YTPDSGQISIALEYMDGGSLADILRMHRTIPEPILSSMFQKLL----RGLSYLHGVRY-- 203
Y D Q + EYM GSL D L EP+ S K+ RGL YLH
Sbjct: 637 YCTDGDQRLLVYEYMPMGSLEDHLFELSHDKEPLNWSTRMKVAVGAARGLEYLHCTADPP 816
Query: 204 LVHRDIKPANLLVNLKGEPKITDFGIS--AGLENSVAMCATFVGTVTYMSPERIRNESYS 261
+++RD+K AN+L++ + PK++DFG++ + ++ + +GT Y +PE + +
Sbjct: 817 VIYRDLKSANILLDNEFNPKLSDFGLAKLGPVGDNTHVSTRVMGTYGYCAPEYAMSGKLT 996
Query: 262 YPADIWSLGLALLESGTG 279
+DI+S G+ LLE TG
Sbjct: 997 LKSDIYSFGVVLLELLTG 1050
>TC17497 UP|CAE45595 (CAE45595) S-receptor kinase-like protein 2, complete
Length = 2946
Score = 70.1 bits (170), Expect = 8e-13
Identities = 68/311 (21%), Positives = 138/311 (43%), Gaps = 26/311 (8%)
Frame = +1
Query: 76 TYRCGSHEMRIFGAIGSGASSVVQRAMHIPTHRVIALKKINIFEKEKRQQLLTEIRTLCE 135
T ++ + +G G V + + + + IA+K+++ + ++ EI+ +
Sbjct: 1648 TISSATNHFSLSNKLGEGGFGPVYKGL-LANGQEIAVKRLSNTSGQGMEEFKNEIKLIAR 1824
Query: 136 APCYEGLVEFHG-AFYTPDSGQISIALEYMDGGSLADILRMHRTIPEPILSSMFQKLLRG 194
+ LV+ G + + ++ + ++ + + + ++ ++ + + + RG
Sbjct: 1825 LQ-HRNLVKLFGCSVHQDENSHANKKMKILLDSTRSKLVDWNKRL------QIIDGIARG 1983
Query: 195 LSYLHGVRYL--VHRDIKPANLLVNLKGEPKITDFGISAGL--ENSVAMCATFVGTVTYM 250
L YLH L +HRD+K +N+L++ + PKI+DFG++ + A +GT YM
Sbjct: 1984 LLYLHQDSRLRIIHRDLKTSNILLDDEMNPKISDFGLARIFIGDQVEARTKRVMGTYGYM 2163
Query: 251 SPERIRNESYSYPADIWSLGLALLESGTGE------------------FPYTANEGPVNL 292
PE + S+S +D++S G+ +LE +G+ + E P+ L
Sbjct: 2164 PPEYAVHGSFSIKSDVFSFGVIVLEIISGKKIGRFYDPHHHLNLLSHAWRLWIEERPLEL 2343
Query: 293 MLQILDDPSPSPSKEKFSPEFCSFVDA---CLQKDPDNRPTAEQLLLHPFITKYETAKVD 349
+ ++LDDP E ++ C+Q+ P+NRP ++L + E K
Sbjct: 2344 VDELLDDP-------VIPTEILRYIHVALLCVQRRPENRPDMLSIVL-MLNGEKELPKPR 2499
Query: 350 LPGFVRSVFDP 360
LP F DP
Sbjct: 2500 LPAFYTGKHDP 2532
>TC8585 weakly similar to PIR|T51339|T51339 mitogen-activated protein
kinase kinase 4 [validated] -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(24%)
Length = 549
Score = 70.1 bits (170), Expect = 8e-13
Identities = 48/101 (47%), Positives = 56/101 (54%), Gaps = 9/101 (8%)
Frame = +2
Query: 251 SPERIRNESYS-----YPADIWSLGLALLESGTGEFPYT-ANEGP--VNLMLQI-LDDPS 301
SPER E+ + ADIWSLGL L E G FP+ A + P V LM I DP
Sbjct: 2 SPERFDPEASGGNYNGFAADIWSLGLTLFELYVGHFPFLQAGQRPDWVTLMCGICFGDPP 181
Query: 302 PSPSKEKFSPEFCSFVDACLQKDPDNRPTAEQLLLHPFITK 342
P E SPEF SFV+ CL+K+ R TA QLL HPF+ K
Sbjct: 182 SLP--ETASPEFRSFVECCLKKESGERWTAAQLLAHPFVCK 298
>TC16248 similar to UP|BAC87845 (BAC87845) Leucine-rich repeat receptor-like
protein kinase 1, partial (17%)
Length = 866
Score = 68.6 bits (166), Expect = 2e-12
Identities = 59/224 (26%), Positives = 105/224 (46%), Gaps = 17/224 (7%)
Frame = +2
Query: 128 TEIRTLCEAPCYEGLVEFHGAFYTPDSGQISIALEYMDGGSLADILRMHRTIPE---PIL 184
+EI+TL E + +++ HG + S + E+++GGSL +L
Sbjct: 2 SEIQTLTEIR-HRNVIKLHG--FCLHSKFSFLVYEFLEGGSLDQVLNSDTQAAAFHWEKR 172
Query: 185 SSMFQKLLRGLSYLHG--VRYLVHRDIKPANLLVNLKGEPKITDFGISAGLENSVAMCAT 242
++ + + LSY+H ++HRDI N+L++L+ E ++DFG + L+
Sbjct: 173 VNVVKGVANALSYMHHDCSPPIIHRDISSKNVLLDLQYEAHVSDFGTAKFLKPGSHTWTA 352
Query: 243 FVGTVTYMSPERIRNESYSYPADIWSLGLALLESGTGEFP-------YTANEGPV--NLM 293
F GT Y +PE + + D++S G+ LE G P + + P+ +L+
Sbjct: 353 FAGTFGYAAPELAQTMQVNEKCDVYSFGVFALEIIMGMHPGDLISSLMSPSTVPMVNDLL 532
Query: 294 LQILDDPSPSPSKEKFSPE---FCSFVDACLQKDPDNRPTAEQL 334
L + D P ++ E S ACL+K+P +RPT +Q+
Sbjct: 533 LIDILDQRPHQVEKPIDEEVILIASLALACLRKNPHSRPTMDQV 664
>TC11528 homologue to UP|Q39868 (Q39868) Protein kinase , partial (56%)
Length = 640
Score = 68.6 bits (166), Expect = 2e-12
Identities = 59/189 (31%), Positives = 90/189 (47%), Gaps = 5/189 (2%)
Frame = +3
Query: 90 IGSGASSVVQRAMHIPTHRVIALKKINIFEK--EKRQQLLTEIRTLCEAPCYEGLVEFHG 147
+GSG V + A T ++A+K I +K E Q+ + R+L + ++ F
Sbjct: 96 LGSGNFGVARLARDKNTGELVAVKYIERGKKIDENVQREIINHRSLR----HPNIIRFKE 263
Query: 148 AFYTPDSGQISIALEYMDGGSLADILRMHRTIPEPILSSMFQKLLRGLSYLHGVRYLVHR 207
TP ++I LEY GG L + + E FQ+L+ G+SY H + + HR
Sbjct: 264 VLLTPT--HLAIVLEYASGGELFERICSAGRFSEDEARYFFQQLISGVSYCHSME-ICHR 434
Query: 208 DIKPANLLVNLKGEP--KITDFGISAGLENSVAMCATFVGTVTYMSPERIRNESY-SYPA 264
D+K N L++ P KI DFG S +T VGT Y++PE + + Y A
Sbjct: 435 DLKLENTLLDGNPSPRLKICDFGYSKSAILHSQPKST-VGTPAYIAPEVLSRKEYDGKVA 611
Query: 265 DIWSLGLAL 273
D+WS G+ L
Sbjct: 612 DVWSCGVTL 638
>TC19452 homologue to UP|Q8S9J9 (Q8S9J9) At1g14000/F7A19_9, partial (41%)
Length = 562
Score = 68.2 bits (165), Expect = 3e-12
Identities = 47/156 (30%), Positives = 79/156 (50%), Gaps = 7/156 (4%)
Frame = +3
Query: 139 YEGLVEFHGAFYTPDSGQISIALEYMDGGSLADILRMHRTIPEPILSSMFQKLLRGLSYL 198
+ +V+F GA + + + EY+ GG L L+ ++ + ++RG++YL
Sbjct: 75 HPNIVQFLGA--VTERKPLMLITEYLRGGDLHQYLKEKGSLSPSTAINFSMDIVRGMAYL 248
Query: 199 HG-VRYLVHRDIKPAN-LLVNLKGEP-KITDFGIS--AGLENS--VAMCATFVGTVTYMS 251
H ++HRD+KP N LLVN + K+ DFG+S ++NS V G+ YM+
Sbjct: 249 HNEPNVIIHRDLKPRNVLLVNSSADHLKVGDFGLSKLITVQNSHDVYKMTGETGSYRYMA 428
Query: 252 PERIRNESYSYPADIWSLGLALLESGTGEFPYTANE 287
PE ++ Y D++S + L E GE P+ + E
Sbjct: 429 PEVFKHRKYDKKVDVYSFAMILYEMLEGEPPFASYE 536
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.319 0.138 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,966,103
Number of Sequences: 28460
Number of extensions: 129214
Number of successful extensions: 855
Number of sequences better than 10.0: 198
Number of HSP's better than 10.0 without gapping: 791
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 795
length of query: 519
length of database: 4,897,600
effective HSP length: 94
effective length of query: 425
effective length of database: 2,222,360
effective search space: 944503000
effective search space used: 944503000
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 57 (26.6 bits)
Medicago: description of AC146560.10


