

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146559.6 + phase: 0
(278 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
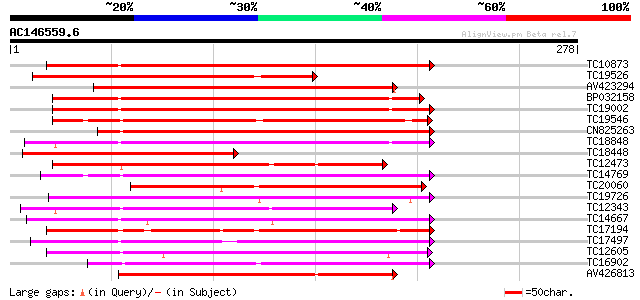
Score E
Sequences producing significant alignments: (bits) Value
TC10873 similar to UP|Q9LDZ5 (Q9LDZ5) F2D10.13 (F5M15.3), partia... 222 6e-59
TC19526 homologue to UP|AAQ96340 (AAQ96340) Protein kinase-like ... 189 4e-49
AV423294 157 2e-39
BP032158 155 9e-39
TC19002 weakly similar to UP|Q9XJ10 (Q9XJ10) ESTs C22458(C62866)... 154 2e-38
TC19546 similar to UP|Q40543 (Q40543) Protein-serine/threonine k... 152 6e-38
CN825263 150 3e-37
TC18848 weakly similar to UP|Q9SSQ6 (Q9SSQ6) F6D8.24 protein, pa... 149 7e-37
TC18448 similar to UP|Q9LZ05 (Q9LZ05) Protein kinase-like, parti... 148 1e-36
TC12473 similar to UP|Q9M068 (Q9M068) Protein kinase-like protei... 147 1e-36
TC14769 UP|Q8LKX1 (Q8LKX1) Receptor-like kinase SYMRK, complete 145 7e-36
TC20060 similar to UP|Q9LZF8 (Q9LZF8) Protein kinase-like (Prote... 138 9e-34
TC19726 similar to UP|Q9XIS3 (Q9XIS3) Lectin-like protein kinase... 138 1e-33
TC12343 similar to UP|CDK9_SCHPO (Q96WV9) Serine/threonine prote... 138 1e-33
TC14667 homologue to UP|Q84P43 (Q84P43) Protein kinase Pti1, com... 136 3e-33
TC17194 UP|CAE02590 (CAE02590) Nod-factor receptor 1b, complete 135 6e-33
TC17497 UP|CAE45595 (CAE45595) S-receptor kinase-like protein 2,... 128 1e-30
TC12605 similar to UP|AAR99871 (AAR99871) Strubbelig receptor fa... 127 3e-30
TC16902 homologue to UP|Q8VZH5 (Q8VZH5) Receptor protein kinase-... 126 4e-30
AV426813 125 1e-29
>TC10873 similar to UP|Q9LDZ5 (Q9LDZ5) F2D10.13 (F5M15.3), partial (77%)
Length = 1478
Score = 222 bits (565), Expect = 6e-59
Identities = 112/190 (58%), Positives = 136/190 (70%)
Frame = +1
Query: 19 AQNFTFRELATATKNFRQECLLSEGGFGRVYKGVIPATGQVVAVKQLDRHGTENSKEFLT 78
A +F FRELA AT+NF++ L+ EGGFG+VYKG + TG+ VAVKQL G + +EF+
Sbjct: 406 AASFGFRELADATRNFKEANLIGEGGFGKVYKGRL-TTGEAVAVKQLSHDGRQGFQEFVM 582
Query: 79 EVSLLSHVHHENLVNLIGYCADGDQRLLVYEYFPGTTLEDRLFENKTDEPPLNWFDRMKV 138
EV +LS +HH NLV LIGYC DGDQRLLVYEY P +LED LFE D+ PLNW RMKV
Sbjct: 583 EVLMLSLLHHTNLVRLIGYCTDGDQRLLVYEYMPMGSLEDHLFELSHDKEPLNWSTRMKV 762
Query: 139 AEAASKGLEYLHDSANPPIIYRDFKAFNILLDVDLNAKLYDFGMVKFSGGDKMNNAPPRV 198
A A++GLEYLH +A+PP+IYRD K+ NILLD + N KL DFG+ K + RV
Sbjct: 763 AVGAARGLEYLHCTADPPVIYRDLKSANILLDNEFNPKLSDFGLAKLGPVGDNTHVSTRV 942
Query: 199 MGTYGYLAPE 208
MGTYGY APE
Sbjct: 943 MGTYGYCAPE 972
>TC19526 homologue to UP|AAQ96340 (AAQ96340) Protein kinase-like protein,
partial (37%)
Length = 432
Score = 189 bits (480), Expect = 4e-49
Identities = 95/140 (67%), Positives = 110/140 (77%)
Frame = +1
Query: 12 GDPTNINAQNFTFRELATATKNFRQECLLSEGGFGRVYKGVIPATGQVVAVKQLDRHGTE 71
G +I AQ FTFRELA ATKNFR ECLL EGGFGRVYKG + +TGQVVAVKQLDR+G +
Sbjct: 22 GPTAHIAAQTFTFRELAAATKNFRPECLLGEGGFGRVYKGRLESTGQVVAVKQLDRNGLQ 201
Query: 72 NSKEFLTEVSLLSHVHHENLVNLIGYCADGDQRLLVYEYFPGTTLEDRLFENKTDEPPLN 131
++EFL EV +LS +HH NLVNLIGYCADGDQRLLVYE+ P +LED L D+ PL+
Sbjct: 202 GNREFLVEVLMLSLLHHPNLVNLIGYCADGDQRLLVYEFMPLGSLEDHL---HGDKEPLD 372
Query: 132 WFDRMKVAEAASKGLEYLHD 151
W RMK+A A+KGLEYLHD
Sbjct: 373 WNTRMKIAAGAAKGLEYLHD 432
>AV423294
Length = 459
Score = 157 bits (396), Expect = 2e-39
Identities = 78/150 (52%), Positives = 101/150 (67%), Gaps = 1/150 (0%)
Frame = +3
Query: 42 EGGFGRVYKGVIPATGQVVAVKQLDRHGTENSKEFLTEVSLLSHVHHENLVNLIGYCADG 101
EGGFG+VY+G I QVVA+KQL+ HG + +EF+ EV LS H NLV LIG+CA+G
Sbjct: 3 EGGFGKVYRGFIERINQVVAIKQLNPHGLQGIREFVVEVLTLSLADHPNLVKLIGFCAEG 182
Query: 102 DQRLLVYEYFPGTTLEDRLFENKTDEPPLNWFDRMKVAEAASKGLEYLHDSANPPIIYRD 161
+QRLLVYEY P +LE L + + PL+W RMK+A A++GLEYLH+ PP+IYRD
Sbjct: 183 EQRLLVYEYMPLGSLESHLHDILPGKKPLDWNTRMKIAAGAARGLEYLHNKMKPPVIYRD 362
Query: 162 FKAFNILLDVDLNAKLYDFGMVKFSG-GDK 190
K N+L+ + KL DFG+ K GDK
Sbjct: 363 IKCSNLLIGDGYHPKLSDFGLAKVGPIGDK 452
>BP032158
Length = 555
Score = 155 bits (391), Expect = 9e-39
Identities = 79/182 (43%), Positives = 112/182 (61%)
Frame = +2
Query: 22 FTFRELATATKNFRQECLLSEGGFGRVYKGVIPATGQVVAVKQLDRHGTENSKEFLTEVS 81
F+ R++ AT NF + EGGFG VYKGV+ + G V+AVKQL + ++EF+ E+
Sbjct: 14 FSLRQIKAATNNFDPANKIGEGGFGPVYKGVL-SEGDVIAVKQLSSKSKQGNREFINEIG 190
Query: 82 LLSHVHHENLVNLIGYCADGDQRLLVYEYFPGTTLEDRLFENKTDEPPLNWFDRMKVAEA 141
++S + H NLV L G C +G+Q LLVYEY +L LF N+ + LNW RMK+
Sbjct: 191 MISALQHPNLVKLYGCCIEGNQLLLVYEYMENNSLARALFGNEEQKLNLNWRTRMKICVG 370
Query: 142 ASKGLEYLHDSANPPIIYRDFKAFNILLDVDLNAKLYDFGMVKFSGGDKMNNAPPRVMGT 201
+KGL YLH+ + I++RD KA N+LLD DL AK+ DFG+ K ++ + R+ GT
Sbjct: 371 IAKGLAYLHEESRLKIVHRDIKATNVLLDKDLKAKISDFGLAKLD-EEENTHISTRIAGT 547
Query: 202 YG 203
G
Sbjct: 548 IG 553
>TC19002 weakly similar to UP|Q9XJ10 (Q9XJ10) ESTs C22458(C62866), partial
(48%)
Length = 780
Score = 154 bits (389), Expect = 2e-38
Identities = 83/187 (44%), Positives = 113/187 (60%)
Frame = +1
Query: 22 FTFRELATATKNFRQECLLSEGGFGRVYKGVIPATGQVVAVKQLDRHGTENSKEFLTEVS 81
F+ +EL +AT NF + L EGGFG VY G + G +AVK+L + EF EV
Sbjct: 28 FSLKELHSATNNFNYDNKLGEGGFGSVYWGQL-WDGSQIAVKRLKVWSNKADMEFAVEVE 204
Query: 82 LLSHVHHENLVNLIGYCADGDQRLLVYEYFPGTTLEDRLFENKTDEPPLNWFDRMKVAEA 141
+L+ V H+NL++L GYCA+G +RL+VY+Y P +L L + E L+W RM +A
Sbjct: 205 ILARVRHKNLLSLRGYCAEGQERLIVYDYMPNLSLLSHLHGQHSSECLLDWNRRMNIAIG 384
Query: 142 ASKGLEYLHDSANPPIIYRDFKAFNILLDVDLNAKLYDFGMVKFSGGDKMNNAPPRVMGT 201
+++G+ YLH A P II+RD KA N+LLD D A++ DFG K D + RV GT
Sbjct: 385 SAEGIVYLHHQATPHIIHRDIKASNVLLDSDFQARVADFGFAKLI-PDGATHVTTRVKGT 561
Query: 202 YGYLAPE 208
GYLAPE
Sbjct: 562 LGYLAPE 582
>TC19546 similar to UP|Q40543 (Q40543) Protein-serine/threonine kinase ,
partial (46%)
Length = 731
Score = 152 bits (384), Expect = 6e-38
Identities = 80/186 (43%), Positives = 119/186 (63%)
Frame = +1
Query: 22 FTFRELATATKNFRQECLLSEGGFGRVYKGVIPATGQVVAVKQLDRHGTENSKEFLTEVS 81
++++++ AT+NF +L +G FG VYK +P TG+VVAVK L + + EF TEV
Sbjct: 199 YSYKDIQKATQNFTT--ILGQGSFGTVYKATMP-TGEVVAVKVLAPNSKQGEHEFQTEVH 369
Query: 82 LLSHVHHENLVNLIGYCADGDQRLLVYEYFPGTTLEDRLFENKTDEPPLNWFDRMKVAEA 141
LL +HH NLVNL+G+C D QR+LVY++ +L + L+ +E L+W +R+++A
Sbjct: 370 LLGRLHHRNLVNLVGFCVDKGQRILVYQFMSNGSLANLLY---GEEKELSWDERLQIAMD 540
Query: 142 ASKGLEYLHDSANPPIIYRDFKAFNILLDVDLNAKLYDFGMVKFSGGDKMNNAPPRVMGT 201
S G+EYLH+ A PP+I+RD K+ NILLD + A + DFG+ K D N+ + GT
Sbjct: 541 ISHGIEYLHEGAVPPVIHRDLKSANILLDDSMRAMVADFGLSKEEIFDGRNSG---LKGT 711
Query: 202 YGYLAP 207
YGY+ P
Sbjct: 712 YGYMDP 729
>CN825263
Length = 663
Score = 150 bits (378), Expect = 3e-37
Identities = 76/165 (46%), Positives = 104/165 (62%)
Frame = +1
Query: 44 GFGRVYKGVIPATGQVVAVKQLDRHGTENSKEFLTEVSLLSHVHHENLVNLIGYCADGDQ 103
GFG VYKG++ G+ VAVK L R +EFL EV +LS +HH NLV LIG C +
Sbjct: 1 GFGLVYKGILN-DGRDVAVKILKRDDQRGGREFLAEVEMLSRLHHRNLVKLIGICIEKQT 177
Query: 104 RLLVYEYFPGTTLEDRLFENKTDEPPLNWFDRMKVAEAASKGLEYLHDSANPPIIYRDFK 163
R L+YE P ++E L + PL+W RMK+A A++GL YLH+ +NP +I+RDFK
Sbjct: 178 RCLIYELVPNGSVESHLHGADKETGPLDWNARMKIALGAARGLAYLHEDSNPCVIHRDFK 357
Query: 164 AFNILLDVDLNAKLYDFGMVKFSGGDKMNNAPPRVMGTYGYLAPE 208
+ NILL+ D K+ DFG+ + + + + VMGT+GYLAPE
Sbjct: 358 SSNILLECDFTPKVSDFGLARTALDEGNKHISTHVMGTFGYLAPE 492
>TC18848 weakly similar to UP|Q9SSQ6 (Q9SSQ6) F6D8.24 protein, partial (64%)
Length = 969
Score = 149 bits (375), Expect = 7e-37
Identities = 85/206 (41%), Positives = 117/206 (56%), Gaps = 5/206 (2%)
Frame = +2
Query: 8 EQMQGDPTNINAQN-----FTFRELATATKNFRQECLLSEGGFGRVYKGVIPATGQVVAV 62
E+++ PT+ + N FT++EL AT F + L EGGFG VY G + G +AV
Sbjct: 209 EKVEEGPTSFGSVNNSWRIFTYKELHAATGGFSDDNKLGEGGFGSVYWGRT-SDGLQIAV 385
Query: 63 KQLDRHGTENSKEFLTEVSLLSHVHHENLVNLIGYCADGDQRLLVYEYFPGTTLEDRLFE 122
K+L ++ EF EV +L V H+NL+ L GYC DQRL+VY+Y P +L L
Sbjct: 386 KKLKAMNSKAEMEFAVEVEVLGRVRHKNLLGLRGYCVGDDQRLIVYDYMPNLSLLSHLHG 565
Query: 123 NKTDEPPLNWFDRMKVAEAASKGLEYLHDSANPPIIYRDFKAFNILLDVDLNAKLYDFGM 182
E LNW RMK+A +++G+ YLH P II+RD KA N+LL+ D + DFG
Sbjct: 566 QFAVEVQLNWQKRMKIAIGSAEGILYLHHEVTPHIIHRDIKASNVLLNSDFEPLVADFGF 745
Query: 183 VKFSGGDKMNNAPPRVMGTYGYLAPE 208
K + +++ RV GT GYLAPE
Sbjct: 746 AKLI-PEGVSHMTTRVKGTLGYLAPE 820
>TC18448 similar to UP|Q9LZ05 (Q9LZ05) Protein kinase-like, partial (30%)
Length = 571
Score = 148 bits (373), Expect = 1e-36
Identities = 70/106 (66%), Positives = 83/106 (78%)
Frame = +1
Query: 7 DEQMQGDPTNINAQNFTFRELATATKNFRQECLLSEGGFGRVYKGVIPATGQVVAVKQLD 66
D G +I AQ F+FR+LATAT+NFR ECLL EGGFGR YKG + + QVVA+KQLD
Sbjct: 241 DASKSGSTDHIAAQTFSFRDLATATRNFRAECLLGEGGFGRAYKGRLESINQVVAIKQLD 420
Query: 67 RHGTENSKEFLTEVSLLSHVHHENLVNLIGYCADGDQRLLVYEYFP 112
R+G + ++EFL EV +LS +HH NLVNLIGYCADGDQRLLVYEY P
Sbjct: 421 RNGLQGNREFLVEVLMLSLLHHPNLVNLIGYCADGDQRLLVYEYMP 558
>TC12473 similar to UP|Q9M068 (Q9M068) Protein kinase-like protein, partial
(42%)
Length = 946
Score = 147 bits (372), Expect = 1e-36
Identities = 77/173 (44%), Positives = 113/173 (64%), Gaps = 9/173 (5%)
Frame = +3
Query: 22 FTFRELATATKNFRQECLLSEGGFGRVYKGVI---------PATGQVVAVKQLDRHGTEN 72
F+F +L +ATK+F+ + L+ EGGFG+VYKG + P +G +VAVK+L+ +
Sbjct: 420 FSFGDLKSATKSFKADALIGEGGFGKVYKGWLDEKKLSPTKPGSGIMVAVKKLNPESMQG 599
Query: 73 SKEFLTEVSLLSHVHHENLVNLIGYCADGDQRLLVYEYFPGTTLEDRLFENKTDEPPLNW 132
E+ +E++ L + H NLV L+GYC D ++ LLVYE+ P +LE+ LF T+ L+W
Sbjct: 600 FHEWQSEINFLGRISHPNLVKLLGYCRDDEEFLLVYEFMPRGSLENHLFRRNTES--LSW 773
Query: 133 FDRMKVAEAASKGLEYLHDSANPPIIYRDFKAFNILLDVDLNAKLYDFGMVKF 185
R+K+A A++GL +LH S +IYRDFKA NILLD + NAK+ FG+ KF
Sbjct: 774 NTRLKIAIGAARGLAFLH-SLXKIVIYRDFKASNILLDGNYNAKISXFGLTKF 929
>TC14769 UP|Q8LKX1 (Q8LKX1) Receptor-like kinase SYMRK, complete
Length = 3495
Score = 145 bits (366), Expect = 7e-36
Identities = 78/193 (40%), Positives = 116/193 (59%)
Frame = +3
Query: 16 NINAQNFTFRELATATKNFRQECLLSEGGFGRVYKGVIPATGQVVAVKQLDRHGTENSKE 75
+++ Q FT + AT+ ++ L+ EGGFG VY+G + GQ VAVK T+ ++E
Sbjct: 2115 SVSIQAFTLEYIEVATERYKT--LIGEGGFGSVYRGTLN-DGQEVAVKVRSSTSTQGTRE 2285
Query: 76 FLTEVSLLSHVHHENLVNLIGYCADGDQRLLVYEYFPGTTLEDRLFENKTDEPPLNWFDR 135
F E++LLS + HENLV L+GYC + DQ++LVY + +L+DRL+ L+W R
Sbjct: 2286 FDNELNLLSAIQHENLVPLLGYCNESDQQILVYPFMSNGSLQDRLYGEPAKRKILDWPTR 2465
Query: 136 MKVAEAASKGLEYLHDSANPPIIYRDFKAFNILLDVDLNAKLYDFGMVKFSGGDKMNNAP 195
+ +A A++GL YLH +I+RD K+ NILLD + AK+ DFG K++ + +
Sbjct: 2466 LSIALGAARGLAYLHTFPGRSVIHRDIKSSNILLDHSMCAKVADFGFSKYAPQEGDSYVS 2645
Query: 196 PRVMGTYGYLAPE 208
V GT GYL PE
Sbjct: 2646 LEVRGTAGYLDPE 2684
>TC20060 similar to UP|Q9LZF8 (Q9LZF8) Protein kinase-like (Protein
serine/threonine kinase), partial (35%)
Length = 482
Score = 138 bits (348), Expect = 9e-34
Identities = 72/149 (48%), Positives = 97/149 (64%), Gaps = 4/149 (2%)
Frame = +1
Query: 60 VAVKQLDRHGTENSKEFLTEVSLLSHVHHENLVNLIGYCADGD----QRLLVYEYFPGTT 115
VAVKQL R G + KE++TEV++L V H NLV L+GYCAD D QRLL+YEY P +
Sbjct: 37 VAVKQLGRRGIQGHKEWVTEVNVLGIVEHPNLVKLVGYCADDDERGIQRLLIYEYMPNRS 216
Query: 116 LEDRLFENKTDEPPLNWFDRMKVAEAASKGLEYLHDSANPPIIYRDFKAFNILLDVDLNA 175
+E L + E PL W R+K+A+ A++GL YLH+ + II+RDFK+ NILLD NA
Sbjct: 217 VEHHL--SPRSETPLPWSRRLKIAQDAARGLTYLHEEMDFQIIFRDFKSSNILLDEHWNA 390
Query: 176 KLYDFGMVKFSGGDKMNNAPPRVMGTYGY 204
KL DFG+ + + + + V+GT GY
Sbjct: 391 KLSDFGLARLGPAEGLTHVSTAVVGTMGY 477
>TC19726 similar to UP|Q9XIS3 (Q9XIS3) Lectin-like protein kinase, partial
(30%)
Length = 728
Score = 138 bits (347), Expect = 1e-33
Identities = 72/192 (37%), Positives = 114/192 (58%), Gaps = 3/192 (1%)
Frame = +3
Query: 20 QNFTFRELATATKNFRQECLLSEGGFGRVYKGVIPATGQVVAVKQLDRHGTENSKEFLTE 79
+ F + EL AT NF ++ L +GG+G VY+G++P VAVK R +++ +FL+E
Sbjct: 126 REFNYVELKKATNNFDEKHKLGQGGYGVVYRGMLPKEKLEVAVKMFSRDKMKSTDDFLSE 305
Query: 80 VSLLSHVHHENLVNLIGYCADGDQRLLVYEYFPGTTLEDRLF--ENKTDEPPLNWFDRMK 137
+ +++ + H++LV L G+C LLVY+Y P +L+ +F E T PL+W R K
Sbjct: 306 LIIINRLRHKHLVRLQGWCHKNGVLLLVYDYMPNGSLDSHIFCEEGGTITTPLSWPLRYK 485
Query: 138 VAEAASKGLEYLHDSANPPIIYRDFKAFNILLDVDLNAKLYDFGMVKFSGGDKMNNAP-P 196
+ + L YLH+ + +++RD KA NI+LD + NAKL DFG+ + +K++
Sbjct: 486 IISGVASALNYLHNEYDQKVVHRDLKASNIMLDSEFNAKLGDFGLARALENEKISYTELE 665
Query: 197 RVMGTYGYLAPE 208
V GT GY+APE
Sbjct: 666 GVHGTMGYIAPE 701
>TC12343 similar to UP|CDK9_SCHPO (Q96WV9) Serine/threonine protein kinase
Cdk9 (Cyclin-dependent kinase Cdk9) , partial (7%)
Length = 723
Score = 138 bits (347), Expect = 1e-33
Identities = 76/188 (40%), Positives = 111/188 (58%), Gaps = 3/188 (1%)
Frame = +3
Query: 6 ADEQMQGDPTNINAQN---FTFRELATATKNFRQECLLSEGGFGRVYKGVIPATGQVVAV 62
++ Q + D NI A+ F++ L ATKNF L EGGFG V+KG + G+ +AV
Sbjct: 150 SEAQNEDDIQNIAAKEHKTFSYETLVAATKNFHAVNKLGEGGFGPVFKGKLN-DGREIAV 326
Query: 63 KQLDRHGTENSKEFLTEVSLLSHVHHENLVNLIGYCADGDQRLLVYEYFPGTTLEDRLFE 122
K+L R + +F+ E LL+ V H N+V+L GYCA G ++LLVYEY P +L+ LF
Sbjct: 327 KKLSRRSNQGRTQFINEAKLLTRVQHRNVVSLFGYCAHGSEKLLVYEYVPRESLDKLLFR 506
Query: 123 NKTDEPPLNWFDRMKVAEAASKGLEYLHDSANPPIIYRDFKAFNILLDVDLNAKLYDFGM 182
++ E L+W R + ++GL YLH+ ++ II+RD KA NILLD K+ DFG+
Sbjct: 507 SQKKE-QLDWKRRFDIISGVARGLLYLHEDSHDCIIHRDIKAANILLDEKWVPKIADFGL 683
Query: 183 VKFSGGDK 190
+ D+
Sbjct: 684 ARIFPEDQ 707
>TC14667 homologue to UP|Q84P43 (Q84P43) Protein kinase Pti1, complete
Length = 1591
Score = 136 bits (343), Expect = 3e-33
Identities = 78/206 (37%), Positives = 115/206 (54%), Gaps = 6/206 (2%)
Frame = +2
Query: 9 QMQGDPTNINAQNFTFRELATATKNFRQECLLSEGGFGRVYKGVIPATGQVVAVKQLD-R 67
+ Q P I + EL T NF + L+ EG +GRVY + G VAVK+LD
Sbjct: 284 ETQKAPPPIEVPALSLDELKEKTDNFGSKALIGEGSYGRVYYATL-NDGNAVAVKKLDVS 460
Query: 68 HGTENSKEFLTEVSLLSHVHHENLVNLIGYCADGDQRLLVYEYFPGTTLEDRLFENKTDE 127
E + EFLT+VS++S + ++N V L GYC +G+ R+L YE+ +L D L K +
Sbjct: 461 SEPETNNEFLTQVSMVSRLKNDNFVELHGYCVEGNLRVLAYEFATMGSLHDILHGRKGVQ 640
Query: 128 -----PPLNWFDRMKVAEAASKGLEYLHDSANPPIIYRDFKAFNILLDVDLNAKLYDFGM 182
P L+W R+++A A++GLEYLH+ P II+RD ++ N+L+ D AK+ DF +
Sbjct: 641 GAQPGPTLDWIQRVRIAVDAARGLEYLHEKVQPAIIHRDIRSSNVLIFEDYKAKIADFNL 820
Query: 183 VKFSGGDKMNNAPPRVMGTYGYLAPE 208
+ RV+GT+GY APE
Sbjct: 821 SNQAPDMAARLHSTRVLGTFGYHAPE 898
>TC17194 UP|CAE02590 (CAE02590) Nod-factor receptor 1b, complete
Length = 2193
Score = 135 bits (341), Expect = 6e-33
Identities = 76/190 (40%), Positives = 115/190 (60%)
Frame = +1
Query: 19 AQNFTFRELATATKNFRQECLLSEGGFGRVYKGVIPATGQVVAVKQLDRHGTENSKEFLT 78
+ F+++ELA AT NF + + +GGFG VY + G+ A+K++D + S EFL
Sbjct: 1042 SMEFSYQELAKATNNFSLDNKIGQGGFGAVYYAEL--RGKKTAIKKMD---VQASTEFLC 1206
Query: 79 EVSLLSHVHHENLVNLIGYCADGDQRLLVYEYFPGTTLEDRLFENKTDEPPLNWFDRMKV 138
E+ +L+HVHH NLV LIGYC +G LVYE+ L L + + + PL W R+++
Sbjct: 1207 ELKVLTHVHHLNLVRLIGYCVEGSL-FLVYEHIDNGNLGQYL--HGSGKEPLPWSSRVQI 1377
Query: 139 AEAASKGLEYLHDSANPPIIYRDFKAFNILLDVDLNAKLYDFGMVKFSGGDKMNNAPPRV 198
A A++GLEY+H+ P I+RD K+ NIL+D +L K+ DFG+ K + R+
Sbjct: 1378 ALDAARGLEYIHEHTVPVYIHRDVKSANILIDKNLRGKVADFGLTKLIEVGN-STLQTRL 1554
Query: 199 MGTYGYLAPE 208
+GT+GY+ PE
Sbjct: 1555 VGTFGYMPPE 1584
>TC17497 UP|CAE45595 (CAE45595) S-receptor kinase-like protein 2, complete
Length = 2946
Score = 128 bits (321), Expect = 1e-30
Identities = 71/198 (35%), Positives = 109/198 (54%)
Frame = +1
Query: 11 QGDPTNINAQNFTFRELATATKNFRQECLLSEGGFGRVYKGVIPATGQVVAVKQLDRHGT 70
+GD A F F +++AT +F L EGGFG VYKG++ A GQ +AVK+L
Sbjct: 1603 RGDEDIDLATIFDFSTISSATNHFSLSNKLGEGGFGPVYKGLL-ANGQEIAVKRLSNTSG 1779
Query: 71 ENSKEFLTEVSLLSHVHHENLVNLIGYCADGDQRLLVYEYFPGTTLEDRLFENKTDEPPL 130
+ +EF E+ L++ + H NLV L G D+ + ++ + T +
Sbjct: 1780 QGMEEFKNEIKLIARLQHRNLVKLFGCSVHQDEN-------SHANKKMKILLDSTRSKLV 1938
Query: 131 NWFDRMKVAEAASKGLEYLHDSANPPIIYRDFKAFNILLDVDLNAKLYDFGMVKFSGGDK 190
+W R+++ + ++GL YLH + II+RD K NILLD ++N K+ DFG+ + GD+
Sbjct: 1939 DWNKRLQIIDGIARGLLYLHQDSRLRIIHRDLKTSNILLDDEMNPKISDFGLARIFIGDQ 2118
Query: 191 MNNAPPRVMGTYGYLAPE 208
+ RVMGTYGY+ PE
Sbjct: 2119 VEARTKRVMGTYGYMPPE 2172
>TC12605 similar to UP|AAR99871 (AAR99871) Strubbelig receptor family 3,
partial (22%)
Length = 680
Score = 127 bits (318), Expect = 3e-30
Identities = 72/192 (37%), Positives = 112/192 (57%), Gaps = 3/192 (1%)
Frame = +3
Query: 19 AQNFTFRELATATKNFRQECLLSEGGFGRVYKGVIPATGQVVAVKQLDRHGTENSK--EF 76
A+ FT L T +F QE L+ G G V K +P G+++AV +LD+ + + K EF
Sbjct: 108 AKCFTIASLQQYTNSFSQENLIGGGMLGSVXKAELP-NGKLLAVXKLDKRASAHQKDDEF 284
Query: 77 LTEVSLLSHVHHENLVNLIGYCADGDQRLLVYEYFPGTTLEDRLFENKTDEPPLNWFDRM 136
L V+ + + H N+V LIGYC++ QRLL+YEY +L D L + + L+W R+
Sbjct: 285 LELVNSIDRIRHANIVELIGYCSEHGQRLLIYEYCSNGSLHDALHSDDEIKTKLSWNARI 464
Query: 137 KVAEAASKGLEYLHDSANPPIIYRDFKAFNILLDVDLNAKLYDFGMVK-FSGGDKMNNAP 195
++A A++ LEYLH+ +PP+I+RD K+ NILLD DL+ + D G+ + D ++
Sbjct: 465 RMALGAARALEYLHELCHPPVIHRDLKSANILLDEDLSVHVSDCGLTPLITFRDL*VSSR 644
Query: 196 PRVMGTYGYLAP 207
++ YGY P
Sbjct: 645 GNLLTAYGYGXP 680
>TC16902 homologue to UP|Q8VZH5 (Q8VZH5) Receptor protein kinase-like
protein, partial (46%)
Length = 941
Score = 126 bits (317), Expect = 4e-30
Identities = 70/170 (41%), Positives = 100/170 (58%)
Frame = +3
Query: 39 LLSEGGFGRVYKGVIPATGQVVAVKQLDRHGTENSKEFLTEVSLLSHVHHENLVNLIGYC 98
LL GGFG+VY G + G VA+K+ + + EF TE+ +LS + H +LV+LIGYC
Sbjct: 12 LLGVGGFGKVYYGEVDG-GTKVAIKRGNPLSEQGVHEFQTEIEMLSKLRHRHLVSLIGYC 188
Query: 99 ADGDQRLLVYEYFPGTTLEDRLFENKTDEPPLNWFDRMKVAEAASKGLEYLHDSANPPII 158
+ + +LVY++ TL + L+ KT +PPL W R+++ A++GL YLH A II
Sbjct: 189 EENTEMILVYDHMAYGTLREHLY--KTQKPPLPWKQRLEICIGAARGLHYLHTGAKYTII 362
Query: 159 YRDFKAFNILLDVDLNAKLYDFGMVKFSGGDKMNNAPPRVMGTYGYLAPE 208
+RD K NILLD AK+ DFG+ K + V G++GYL PE
Sbjct: 363 HRDVKTTNILLDEKWVAKVSDFGLSKTGPTLDNTHVSTVVKGSFGYLDPE 512
>AV426813
Length = 431
Score = 125 bits (313), Expect = 1e-29
Identities = 66/138 (47%), Positives = 91/138 (65%), Gaps = 1/138 (0%)
Frame = +3
Query: 54 PATGQVVAVKQLDRHGTENSKEFLTEVSLLSHVHHENLVNLIGYCADGDQRLLVYEYFPG 113
P TG V+AVK+L++ G++ E+LTE++ L + H NLV LIGY + D R+LVYE+
Sbjct: 15 PGTGFVIAVKRLNQDGSQGHSEWLTEINYLGQLRHPNLVKLIGYSIEDDHRILVYEFLAK 194
Query: 114 TTLEDRLFENKTDEPPLNWFDRMKVAEAASKGLEYLHDSANPPIIYRDFKAFNILLDVDL 173
+L++ LF + PL+W RMK+A A+KGL +LH S +IYRDFK NIL+D +
Sbjct: 195 GSLDNHLFRRASYFQPLSWNIRMKIALDAAKGLAFLH-SDEVEVIYRDFKTSNILIDSNY 371
Query: 174 NAKLYDFGMVKFS-GGDK 190
NAKL DFG+ K GDK
Sbjct: 372 NAKLSDFGLAKDGPAGDK 425
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.319 0.137 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,787,092
Number of Sequences: 28460
Number of extensions: 41144
Number of successful extensions: 449
Number of sequences better than 10.0: 189
Number of HSP's better than 10.0 without gapping: 340
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 342
length of query: 278
length of database: 4,897,600
effective HSP length: 89
effective length of query: 189
effective length of database: 2,364,660
effective search space: 446920740
effective search space used: 446920740
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 55 (25.8 bits)
Medicago: description of AC146559.6


