

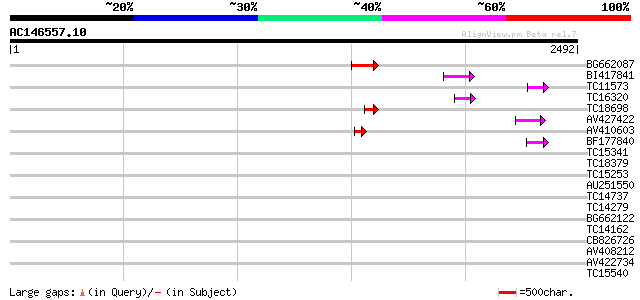
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146557.10 - phase: 0
(2492 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BG662087 135 1e-31
BI417841 72 1e-12
TC11573 similar to UP|Q9M6N4 (Q9M6N4) Pol protein integrase regi... 63 6e-10
TC16320 weakly similar to UP|Q84TE0 (Q84TE0) At5g51080, partial ... 59 1e-08
TC18698 55 1e-07
AV427422 55 1e-07
AV410603 53 7e-07
BF177840 48 2e-05
TC15341 similar to UP|Q9M0H8 (Q9M0H8) Predicted proline-rich pro... 35 0.14
TC18379 similar to UP|Q9SBM1 (Q9SBM1) Hydroxyproline-rich glycop... 35 0.14
TC15253 weakly similar to UP|Q9ZT16 (Q9ZT16) Arabinogalactan-pro... 34 0.24
AU251550 34 0.24
TC14737 similar to UP|Q9LEB3 (Q9LEB3) RNA binding protein 47, pa... 33 0.41
TC14279 similar to UP|Q09085 (Q09085) Hydroxyproline-rich glycop... 33 0.70
BG662122 32 0.92
TC14162 similar to UP|OLP1_LYCES (Q41350) Osmotin-like protein p... 32 0.92
CB826726 32 0.92
AV408212 32 0.92
AV422734 32 1.2
TC15540 similar to UP|Q9NGS5 (Q9NGS5) Prespore protein MF12 (Fra... 32 1.6
>BG662087
Length = 373
Score = 135 bits (339), Expect = 1e-31
Identities = 64/119 (53%), Positives = 84/119 (69%)
Frame = +1
Query: 1500 GKVRMCVDFRDLNKASPKDNFPLPHIDVLVDNTAQSKVFSFMDGFSGYNQIKMSPEDREK 1559
GK RM VD+ DLNKA PKD++PLP ID LVD + +++ S MD +SGY+QIKM P D +K
Sbjct: 16 GKWRMWVDYTDLNKACPKDSYPLPSIDKLVDGASDNELLSLMDAYSGYHQIKMHPSDEDK 195
Query: 1560 TSFITPWGTFCYKVMPFGLINAGATYQRGMTTLFHDMTHKEVEVYVDDMIVKSTDEEQH 1618
T+F+T +CY+ +PFGL NAGATYQ M +F D + +EVY+D+MIVKS H
Sbjct: 196 TAFMTARVNYCYQTIPFGLKNAGATYQXLMDRVFXDXVGRNMEVYLDNMIVKSALRANH 372
>BI417841
Length = 617
Score = 71.6 bits (174), Expect = 1e-12
Identities = 41/136 (30%), Positives = 66/136 (48%), Gaps = 2/136 (1%)
Frame = +1
Query: 1907 NSKWGLVFDGAV--NAYGKGIGAVIVSPQGHHIPFTARILFECTNNMAEYEACIFGIEEA 1964
N L FDG+ N G GAV+ + G + + + TNN AEY I G++ A
Sbjct: 127 NRSCTLEFDGSSKGNPGSAGAGAVLRAEDGSKV-YLREGVGNQTNNQAEYRGLILGLKHA 303
Query: 1965 IDMRIKHLDIYGDSALVINQIKGEWETHHAKLIPYRDYARRLLTYFTKVELHHIPRDENQ 2024
+ +H+++ GDS LV Q++G W+ + + + A+ L + F +++H+PR N
Sbjct: 304 HEQGYQHINVKGDSQLVCKQVEGSWKARNPNIASLCNEAKELKSKFQSFDINHVPRQYNS 483
Query: 2025 MADALATLSSMFRVNH 2040
AD A L H
Sbjct: 484 EADVQANLGVNLPAGH 531
>TC11573 similar to UP|Q9M6N4 (Q9M6N4) Pol protein integrase region
(Fragment), partial (10%)
Length = 572
Score = 62.8 bits (151), Expect = 6e-10
Identities = 36/96 (37%), Positives = 49/96 (50%), Gaps = 1/96 (1%)
Frame = +2
Query: 2274 DNGTNLNNNVVQALCEEFKIEHHNSSPYRPQMNGAVEAANKNI-KRIVQKMVTTYKDWHE 2332
+NG + Q C+ I+ SS PQ NG EAANK I K I +++ W +
Sbjct: 8 ENGIQFTSKQTQDFCDGMGIQMRFSSVKHPQTNGQTEAANKVILKGIKRRLYEAEGRWID 187
Query: 2333 MLPYALHGYRTTVRSSTGATPFSLVYGMEAVLPLEV 2368
LP L Y T +SS TPF L YG + +LP+E+
Sbjct: 188 ELPIVLWSYNTMPQSSIKETPF*LTYGADTMLPVEI 295
>TC16320 weakly similar to UP|Q84TE0 (Q84TE0) At5g51080, partial (11%)
Length = 632
Score = 58.5 bits (140), Expect = 1e-08
Identities = 29/91 (31%), Positives = 44/91 (47%)
Frame = +2
Query: 1954 YEACIFGIEEAIDMRIKHLDIYGDSALVINQIKGEWETHHAKLIPYRDYARRLLTYFTKV 2013
Y I G++ AI KH+ + GDS LV NQ++G W+ + + A+ L F
Sbjct: 2 YRGLILGLKHAIKEGYKHIQVKGDSMLVCNQVQGLWKIKNQNIASLCSEAKELKNKFLSF 181
Query: 2014 ELHHIPRDENQMADALATLSSMFRVNHWNDV 2044
+++HIPR+ N AD A R +V
Sbjct: 182 KINHIPREYNSEADVQANFGISLRAGQVEEV 274
>TC18698
Length = 808
Score = 55.5 bits (132), Expect = 1e-07
Identities = 30/62 (48%), Positives = 38/62 (60%)
Frame = -2
Query: 1557 REKTSFITPWGTFCYKVMPFGLINAGATYQRGMTTLFHDMTHKEVEVYVDDMIVKSTDEE 1616
++KT+ + Y+VMP GL N TYQR M +FH K VEVYV+DMIVKS+ E
Sbjct: 807 KKKTTLKINRVNYYYQVMPLGLKNI*TTYQRLMDKIFHKQI*KNVEVYVEDMIVKSSQE* 628
Query: 1617 QH 1618
H
Sbjct: 627 FH 622
>AV427422
Length = 417
Score = 55.1 bits (131), Expect = 1e-07
Identities = 33/133 (24%), Positives = 63/133 (46%), Gaps = 2/133 (1%)
Frame = +1
Query: 2223 SNGHRFILVAIDYFTKWVEAASYTN-VTKQVVAKFIKNNIICRYGVPSKIITDNGTNLNN 2281
S G+ +LV +D +K+ + T +V+A ++ +GVP I++D +
Sbjct: 19 SKGYEAVLVVVDRLSKFSHFVPLKHPYTAKVIADIFVREVVRLHGVPLSIVSDRDPLFMS 198
Query: 2282 NVVQALCEEFKIEHHNSSPYRPQMNGAVEAANKNIKRIVQKMVTTY-KDWHEMLPYALHG 2340
N + L + + S+ Y P+ +G E N+ ++ ++ + K W +P+A +
Sbjct: 199 NFWKELFKMQGTKLKMSTAYHPESDGQTEVVNRCLETYLRCFIADQPKSWAHWVPWAEYW 378
Query: 2341 YRTTVRSSTGATP 2353
Y T+ STG TP
Sbjct: 379 YNTSYHVSTGQTP 417
>AV410603
Length = 162
Score = 52.8 bits (125), Expect = 7e-07
Identities = 26/51 (50%), Positives = 34/51 (65%)
Frame = +1
Query: 1517 KDNFPLPHIDVLVDNTAQSKVFSFMDGFSGYNQIKMSPEDREKTSFITPWG 1567
KD+FP+P +D L+D S+ FS +D SGY+QI + PEDR KT F T G
Sbjct: 10 KDSFPMPTVDELLDELRGSQFFSKLDLRSGYHQILVKPEDRHKTVFRTHHG 162
>BF177840
Length = 410
Score = 48.1 bits (113), Expect = 2e-05
Identities = 26/101 (25%), Positives = 49/101 (47%), Gaps = 1/101 (0%)
Frame = +2
Query: 2269 SKIITDNGTNLNNNVVQALCEEFKIEHHNSSPYRPQMNGAVEAANKNIKRIVQKMVT-TY 2327
+ I++D T ++ + L + + S+ PQ +G E NK + +++ ++
Sbjct: 11 TSIVSDRDTKFISHFWRTLWGKVGTKLLYSTTCHPQTDGQTEVVNKTLSTLLRSVLERNL 190
Query: 2328 KDWHEMLPYALHGYRTTVRSSTGATPFSLVYGMEAVLPLEV 2368
K W LP+ Y V S+T +PF +VYG + PL++
Sbjct: 191 KMWETWLPHIEFAYNRVVHSTTKHSPFEIVYGYNPLTPLDL 313
>TC15341 similar to UP|Q9M0H8 (Q9M0H8) Predicted proline-rich protein,
partial (33%)
Length = 1002
Score = 35.0 bits (79), Expect = 0.14
Identities = 40/141 (28%), Positives = 54/141 (37%), Gaps = 26/141 (18%)
Frame = +1
Query: 622 AGQAMATVAPINAAQMPPSYPYMPYSQHPFFPPFYHQYP----LPPGQ----PQV----- 668
A Q A + A P+Y Y+P + P Q P LPP Q PQ+
Sbjct: 607 AAQPQAPAPNVTQAPQQPAY-YIPSTPLPGNTQTVAQLPQNQYLPPDQQYRTPQMQDMSR 783
Query: 669 -------------PVNAIAQQMKQQLPVQQQQQQQQQQYTRPTFPPIPMLYAELLPTLLQ 715
P + Q + Q P QQQQQQQQQQ P ++ PT Q
Sbjct: 784 VAPQPCTNXSGXSPPPPVQQYPQYQQPQQQQQQQQQQQQWPQQLP------QQVQPT--Q 939
Query: 716 RGHCTTRQGKPPPDPLPPRFR 736
+ Q +PP P+ P ++
Sbjct: 940 PPSMQSPQIRPPSSPVYPPYQ 1002
>TC18379 similar to UP|Q9SBM1 (Q9SBM1) Hydroxyproline-rich glycoprotein
DZ-HRGP precursor, partial (8%)
Length = 584
Score = 35.0 bits (79), Expect = 0.14
Identities = 27/94 (28%), Positives = 34/94 (35%), Gaps = 4/94 (4%)
Frame = +1
Query: 630 APINAAQMPPSYPYMPYSQHPFFPPFYHQYPLPPGQPQVPVNAIAQQMKQQLPVQQQQQQ 689
AP + P YP +PY QH F+PP P PP P P P
Sbjct: 172 APPYGMEFSP-YPSLPYPQHQFYPPPMAPPPPPPPAPAPPA-----------PPPYWAAP 315
Query: 690 QQQQYTRP----TFPPIPMLYAELLPTLLQRGHC 719
+ Y P T PP P A + ++ HC
Sbjct: 316 PPRPYGPPLFTSTLPPRPPKPAPPKNVVKEKSHC 417
>TC15253 weakly similar to UP|Q9ZT16 (Q9ZT16) Arabinogalactan-protein
(ATAGP4) (AT5G10430/F12B17_220), partial (42%)
Length = 575
Score = 34.3 bits (77), Expect = 0.24
Identities = 19/74 (25%), Positives = 29/74 (38%), Gaps = 2/74 (2%)
Frame = +1
Query: 639 PSYPYMPYSQHPFFPPFYHQYP--LPPGQPQVPVNAIAQQMKQQLPVQQQQQQQQQQYTR 696
P + + Q P PP HQ P LPP Q+P+ + + LP+ Q +
Sbjct: 217 PHLQQLHHQQLPLLPPLLHQLPPLLPPPPLQLPLQPLPPPVHPPLPLPLHPPQSHPAQHQ 396
Query: 697 PTFPPIPMLYAELL 710
PP+ L+
Sbjct: 397 AQAPPLDQAQTALI 438
>AU251550
Length = 329
Score = 34.3 bits (77), Expect = 0.24
Identities = 18/40 (45%), Positives = 21/40 (52%), Gaps = 1/40 (2%)
Frame = +2
Query: 638 PPSYPYMPYSQHPFFPPFYHQYPLPP-GQPQVPVNAIAQQ 676
PP P PY+ P PPF H +P PP G P P A+ Q
Sbjct: 140 PPGPPRDPYAPAPPPPPFGHHHPPPPHGPPSPPPPAMFYQ 259
>TC14737 similar to UP|Q9LEB3 (Q9LEB3) RNA binding protein 47, partial (32%)
Length = 803
Score = 33.5 bits (75), Expect = 0.41
Identities = 27/72 (37%), Positives = 32/72 (43%), Gaps = 4/72 (5%)
Frame = +3
Query: 624 QAMATVAPINAAQMPPSYPYMPYSQHPFFPPFYHQYPLPPGQPQ----VPVNAIAQQMKQ 679
Q MA P A M M Y QH + P+ H + P Q Q A+ QQ Q
Sbjct: 213 QWMAMQYPATAMAMMQQQMMM-YPQH--YMPYVHHHYQPHHQQQQAAAAAAGAVPQQQHQ 383
Query: 680 QLPVQQQQQQQQ 691
Q QQQQ+QQQ
Sbjct: 384 QHHHQQQQKQQQ 419
>TC14279 similar to UP|Q09085 (Q09085) Hydroxyproline-rich glycoprotein
(HRGP) (Fragment), partial (57%)
Length = 941
Score = 32.7 bits (73), Expect = 0.70
Identities = 26/99 (26%), Positives = 32/99 (32%), Gaps = 4/99 (4%)
Frame = +1
Query: 638 PPSYPYMPYSQHPFFPPFYHQYPLPPGQPQVPVNAIAQQMKQQLPVQQQQQQQQQQYTRP 697
PP P Y + P PP+Y++ P PP P Q P Y P
Sbjct: 31 PPPVPKPYYYKSPPPPPYYYKSPPPPSPSPPP-----PYYYQSPPPPSHSPPPPYYYKSP 195
Query: 698 ----TFPPIPMLYAELLPTLLQRGHCTTRQGKPPPDPLP 732
PP P Y P + PPP P+P
Sbjct: 196 PPPSPSPPPPYYYKSPPPPSPSPPPPYYYKSPPPPSPIP 312
>BG662122
Length = 386
Score = 32.3 bits (72), Expect = 0.92
Identities = 18/60 (30%), Positives = 32/60 (53%), Gaps = 2/60 (3%)
Frame = +2
Query: 714 LQRGHCTTRQGKPPPDPLPPRFRSDLK--CDFHQGALGHDVEGCYALKYIVKKLIDQGKL 771
++ H + G+ P PPR D C++ + + HD++ C+ LK ++KLI G+L
Sbjct: 134 VKAAHMVGKSGQS*P---PPRRGIDTTK*CEYRRSVV-HDIDDCFTLKREIEKLIKMGRL 301
>TC14162 similar to UP|OLP1_LYCES (Q41350) Osmotin-like protein precursor,
partial (87%)
Length = 1232
Score = 32.3 bits (72), Expect = 0.92
Identities = 18/71 (25%), Positives = 34/71 (47%)
Frame = +3
Query: 633 NAAQMPPSYPYMPYSQHPFFPPFYHQYPLPPGQPQVPVNAIAQQMKQQLPVQQQQQQQQQ 692
N + S+ +P+S H PP PLP + Q+ ++ +A+ Q+ P ++++ +
Sbjct: 30 NGSSFFTSHILLPHSHH-HHPPHTFTCPLPNPRQQLQIHRLARHPAQRRPPSPRRRRLRP 206
Query: 693 QYTRPTFPPIP 703
Q P P P
Sbjct: 207 QPLHPPLHPRP 239
>CB826726
Length = 505
Score = 32.3 bits (72), Expect = 0.92
Identities = 19/49 (38%), Positives = 25/49 (50%), Gaps = 9/49 (18%)
Frame = +2
Query: 657 HQYPLPPGQPQV---------PVNAIAQQMKQQLPVQQQQQQQQQQYTR 696
++YPL P P + P Q + +LP QQQQQQQQQ Y +
Sbjct: 275 NRYPLHPPPPGIGYGRGGGPNPNPNPNQGFQPRLPYQQQQQQQQQHYVQ 421
>AV408212
Length = 362
Score = 32.3 bits (72), Expect = 0.92
Identities = 15/26 (57%), Positives = 18/26 (68%)
Frame = +3
Query: 672 AIAQQMKQQLPVQQQQQQQQQQYTRP 697
A+ QQ +QQ QQQQQQQQ Q +P
Sbjct: 84 ALQQQQQQQQQQQQQQQQQQSQQQQP 161
Score = 29.3 bits (64), Expect = 7.8
Identities = 14/22 (63%), Positives = 16/22 (72%)
Frame = +3
Query: 671 NAIAQQMKQQLPVQQQQQQQQQ 692
+A QQ +QQ QQQQQQQQQ
Sbjct: 78 HAALQQQQQQQQQQQQQQQQQQ 143
>AV422734
Length = 448
Score = 32.0 bits (71), Expect = 1.2
Identities = 26/72 (36%), Positives = 30/72 (41%)
Frame = +1
Query: 629 VAPINAAQMPPSYPYMPYSQHPFFPPFYHQYPLPPGQPQVPVNAIAQQMKQQLPVQQQQQ 688
+ PIN Q P P P HQ P N + QQ +QQ P QQQ
Sbjct: 277 IRPINLQQNPNPNPNNPILN-------LHQNPNS--------NHLQQQQQQQQP---QQQ 402
Query: 689 QQQQQYTRPTFP 700
QQQQ+ RP P
Sbjct: 403 QQQQKMVRPGNP 438
>TC15540 similar to UP|Q9NGS5 (Q9NGS5) Prespore protein MF12 (Fragment),
partial (6%)
Length = 806
Score = 31.6 bits (70), Expect = 1.6
Identities = 19/51 (37%), Positives = 26/51 (50%)
Frame = +2
Query: 666 PQVPVNAIAQQMKQQLPVQQQQQQQQQQYTRPTFPPIPMLYAELLPTLLQR 716
PQV ++Q +QQLP QQQQ Q Q + P + L + LP Q+
Sbjct: 92 PQVQQQQLSQMQQQQLPQLQQQQLSQLQ-PQQQLPQLQQLQHQQLPQQQQQ 241
Score = 30.0 bits (66), Expect = 4.5
Identities = 19/51 (37%), Positives = 28/51 (54%), Gaps = 1/51 (1%)
Frame = +2
Query: 667 QVPVNAIAQQMKQQLP-VQQQQQQQQQQYTRPTFPPIPMLYAELLPTLLQR 716
Q+P QQ++QQ P + Q QQQQQQQ + + + + LP L Q+
Sbjct: 5 QLPQLQQQQQLQQQPPQLTQLQQQQQQQLPQVQQQQLSQMQQQQLPQLQQQ 157
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.319 0.136 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 42,319,216
Number of Sequences: 28460
Number of extensions: 611742
Number of successful extensions: 4030
Number of sequences better than 10.0: 69
Number of HSP's better than 10.0 without gapping: 3541
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3851
length of query: 2492
length of database: 4,897,600
effective HSP length: 106
effective length of query: 2386
effective length of database: 1,880,840
effective search space: 4487684240
effective search space used: 4487684240
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 63 (28.9 bits)
Medicago: description of AC146557.10


