

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146554.3 - phase: 0
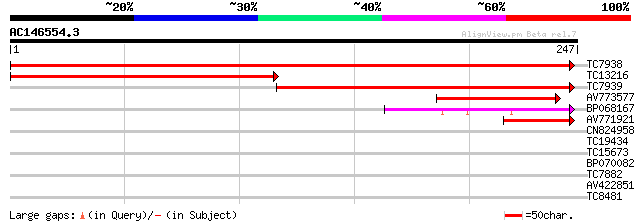
(247 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC7938 homologue to UP|Q8LJW0 (Q8LJW0) 40S ribosomal S4 protein,... 456 e-129
TC13216 UP|Q8LJW0 (Q8LJW0) 40S ribosomal S4 protein, partial (51%) 237 1e-63
TC7939 similar to UP|RS4_PRUAR (O81363) 40S ribosomal protein S4... 235 5e-63
AV773577 87 2e-18
BP068167 60 3e-10
AV771921 43 6e-05
CN824958 28 1.1
TC19434 similar to UP|Q94F63 (Q94F63) Somatic embryogenesis rece... 27 2.5
TC15673 similar to UP|Q8LG90 (Q8LG90) Dihydrodipicolinate reduct... 26 7.3
BP070082 26 7.3
TC7882 homologue to UP|O49875 (O49875) Adenine nucleotide transl... 26 7.3
AV422851 25 9.5
TC8481 similar to UP|Q9XEY0 (Q9XEY0) Nt-gh3 deduced protein, par... 25 9.5
>TC7938 homologue to UP|Q8LJW0 (Q8LJW0) 40S ribosomal S4 protein, complete
Length = 1142
Score = 456 bits (1173), Expect = e-129
Identities = 220/246 (89%), Positives = 236/246 (95%)
Frame = +2
Query: 1 MLDKLGGAFAPKPSSGPHKSRECLPLILILRNRLKYALTYREVIAILMQRHVLVDGKVRT 60
MLDKLGGAFAPKPSSGPHKSRECLPLILILRNRLKYALTYREVIAILMQRH+LVDGKVRT
Sbjct: 104 MLDKLGGAFAPKPSSGPHKSRECLPLILILRNRLKYALTYREVIAILMQRHILVDGKVRT 283
Query: 61 DKTYPAGFMDVVSIPKTNENFRLLYDTKGRFRLHSVRDDEAKFKLCKVRSVQFGQKGIPY 120
DKTYP+GFMDVVSIPKTNENFRLLYDTKGRFRLHSVRDDEAKFKLCKVRSVQFGQKGIPY
Sbjct: 284 DKTYPSGFMDVVSIPKTNENFRLLYDTKGRFRLHSVRDDEAKFKLCKVRSVQFGQKGIPY 463
Query: 121 LNTYDGRTIRYPDPLIKANDTIKLDLEQNKITDFIKFDVGNVVMVTGGRNRGRVGFIKNR 180
LNTYDGRTIRYPDP+I+ANDTIKLDL++NKI DFIKFDVGNVVMVTGGRNRGRVG IK+R
Sbjct: 464 LNTYDGRTIRYPDPVIRANDTIKLDLQENKIVDFIKFDVGNVVMVTGGRNRGRVGVIKSR 643
Query: 181 KKHMGTFETIHVQDAAGHEFATRLGNVFTIGRGPKPWVSLPKGRGIKLTVIEEARKRMAA 240
+ G+F+TIHVQDA GHEFATRLGNVFTIG+G KPW+SLPKG+GIKL++IEEARKR+AA
Sbjct: 644 DRQKGSFDTIHVQDATGHEFATRLGNVFTIGKGSKPWISLPKGKGIKLSIIEEARKRIAA 823
Query: 241 SQQAAA 246
Q AA
Sbjct: 824 QQATAA 841
>TC13216 UP|Q8LJW0 (Q8LJW0) 40S ribosomal S4 protein, partial (51%)
Length = 450
Score = 237 bits (605), Expect = 1e-63
Identities = 116/117 (99%), Positives = 117/117 (99%)
Frame = +1
Query: 1 MLDKLGGAFAPKPSSGPHKSRECLPLILILRNRLKYALTYREVIAILMQRHVLVDGKVRT 60
MLDKLGGAFAPKPSSGPHKSRECLPLILILRNRLKYALTYREVIAILMQRH+LVDGKVRT
Sbjct: 100 MLDKLGGAFAPKPSSGPHKSRECLPLILILRNRLKYALTYREVIAILMQRHILVDGKVRT 279
Query: 61 DKTYPAGFMDVVSIPKTNENFRLLYDTKGRFRLHSVRDDEAKFKLCKVRSVQFGQKG 117
DKTYPAGFMDVVSIPKTNENFRLLYDTKGRFRLHSVRDDEAKFKLCKVRSVQFGQKG
Sbjct: 280 DKTYPAGFMDVVSIPKTNENFRLLYDTKGRFRLHSVRDDEAKFKLCKVRSVQFGQKG 450
>TC7939 similar to UP|RS4_PRUAR (O81363) 40S ribosomal protein S4, partial
(49%)
Length = 651
Score = 235 bits (600), Expect = 5e-63
Identities = 114/130 (87%), Positives = 121/130 (92%)
Frame = +1
Query: 117 GIPYLNTYDGRTIRYPDPLIKANDTIKLDLEQNKITDFIKFDVGNVVMVTGGRNRGRVGF 176
GIPYLNTYDGRTIRYPDPLIKANDTIKLDLE NKIT+FIKFDVGNVVMVTGGRNRGRVG
Sbjct: 1 GIPYLNTYDGRTIRYPDPLIKANDTIKLDLELNKITEFIKFDVGNVVMVTGGRNRGRVGV 180
Query: 177 IKNRKKHMGTFETIHVQDAAGHEFATRLGNVFTIGRGPKPWVSLPKGRGIKLTVIEEARK 236
IKNR+K G+F+TIHVQDA GHEFATRLGNVF IG+G KPWVSLPKGRGIKL++IEEARK
Sbjct: 181 IKNREKQKGSFDTIHVQDATGHEFATRLGNVFMIGKGSKPWVSLPKGRGIKLSIIEEARK 360
Query: 237 RMAASQQAAA 246
R AA QQAAA
Sbjct: 361 RQAAQQQAAA 390
>AV773577
Length = 452
Score = 87.4 bits (215), Expect = 2e-18
Identities = 42/54 (77%), Positives = 47/54 (86%)
Frame = -1
Query: 187 FETIHVQDAAGHEFATRLGNVFTIGRGPKPWVSLPKGRGIKLTVIEEARKRMAA 240
F+TI V DA GHEFATRLGNVF IG+G KPWVSLP GRGI+L++IEEARKR AA
Sbjct: 452 FDTIPVPDATGHEFATRLGNVFMIGQGSKPWVSLP*GRGIQLSIIEEARKRPAA 291
>BP068167
Length = 429
Score = 60.1 bits (144), Expect = 3e-10
Identities = 37/86 (43%), Positives = 48/86 (55%), Gaps = 3/86 (3%)
Frame = -1
Query: 164 MVTGGRNRGRVGFIKNRKKHMGTF-ETIHVQDAAGH-EFATRLGNVFTIGRGPKPW-VSL 220
MVTGGRNRGRVG I+ R + G DA GH +RLGNVFTIG+G +PW +S
Sbjct: 429 MVTGGRNRGRVGVIQTRDRQQGKL*HNPLSTDATGHSSLLSRLGNVFTIGQGSEPWTISS 250
Query: 221 PKGRGIKLTVIEEARKRMAASQQAAA 246
R + + + + + QQ AA
Sbjct: 249 QALRVLSYQSLRKLGRGLLPDQQPAA 172
>AV771921
Length = 451
Score = 42.7 bits (99), Expect = 6e-05
Identities = 21/31 (67%), Positives = 24/31 (76%)
Frame = -2
Query: 216 PWVSLPKGRGIKLTVIEEARKRMAASQQAAA 246
PWVSLP RGIKL+++ EARKR AA AAA
Sbjct: 450 PWVSLP*ARGIKLSILVEARKRPAALPPAAA 358
>CN824958
Length = 593
Score = 28.5 bits (62), Expect = 1.1
Identities = 16/38 (42%), Positives = 21/38 (55%)
Frame = +2
Query: 1 MLDKLGGAFAPKPSSGPHKSRECLPLILILRNRLKYAL 38
ML +LGG+ P+PSS RE P L+ RL + L
Sbjct: 176 MLTRLGGSLVPQPSSLQRVRRETRPRSDHLQRRLCFLL 289
>TC19434 similar to UP|Q94F63 (Q94F63) Somatic embryogenesis receptor-like
kinase 2, partial (26%)
Length = 550
Score = 27.3 bits (59), Expect = 2.5
Identities = 18/53 (33%), Positives = 25/53 (46%), Gaps = 2/53 (3%)
Frame = +2
Query: 112 QFGQ-KGIPYLNTYDGR-TIRYPDPLIKANDTIKLDLEQNKITDFIKFDVGNV 162
Q GQ + YL Y T + PD L + + LDL N +T I +GN+
Sbjct: 29 QLGQLSNLQYLELYSNNMTGKIPDELGNLTNLVSLDLYLNNLTGNIPSTLGNL 187
>TC15673 similar to UP|Q8LG90 (Q8LG90) Dihydrodipicolinate reductase-like
protein, partial (10%)
Length = 682
Score = 25.8 bits (55), Expect = 7.3
Identities = 11/35 (31%), Positives = 20/35 (56%)
Frame = +2
Query: 200 FATRLGNVFTIGRGPKPWVSLPKGRGIKLTVIEEA 234
F T +G + + + W+S P G+G + ++EEA
Sbjct: 503 FQTWIGYYWYVVSTFQTWISFPNGQGTQPFIMEEA 607
>BP070082
Length = 489
Score = 25.8 bits (55), Expect = 7.3
Identities = 20/59 (33%), Positives = 28/59 (46%), Gaps = 2/59 (3%)
Frame = +1
Query: 8 AFAPKPSSGPHKSRECLPLILILRNRLKYALTYREVIAI--LMQRHVLVDGKVRTDKTY 64
A +PSS P S L L + L ++K + +AI L+Q+ KVR D TY
Sbjct: 136 ALGSRPSSSPSSSSS*LALRIKLWKKVKIDVIMANNLAILNLLQKSNS*KVKVRIDDTY 312
>TC7882 homologue to UP|O49875 (O49875) Adenine nucleotide translocator,
complete
Length = 1656
Score = 25.8 bits (55), Expect = 7.3
Identities = 13/65 (20%), Positives = 27/65 (41%)
Frame = +2
Query: 177 IKNRKKHMGTFETIHVQDAAGHEFATRLGNVFTIGRGPKPWVSLPKGRGIKLTVIEEARK 236
I R KH ++H+ + F +F +G PW+ I+ + ++ R+
Sbjct: 71 ISLRSKHAAHLNSVHISSCSSPSFTPSPPFLFDSEKGKAPWL-------IRFSTLQSWRR 229
Query: 237 RMAAS 241
+ +S
Sbjct: 230 LLGSS 244
>AV422851
Length = 419
Score = 25.4 bits (54), Expect = 9.5
Identities = 11/22 (50%), Positives = 15/22 (68%)
Frame = +2
Query: 74 IPKTNENFRLLYDTKGRFRLHS 95
+PK +N +LL +KGR LHS
Sbjct: 134 MPKGYQNLQLLLKSKGRTMLHS 199
>TC8481 similar to UP|Q9XEY0 (Q9XEY0) Nt-gh3 deduced protein, partial (76%)
Length = 1713
Score = 25.4 bits (54), Expect = 9.5
Identities = 12/21 (57%), Positives = 15/21 (71%)
Frame = +3
Query: 202 TRLGNVFTIGRGPKPWVSLPK 222
TRL NV T+G PW+SLP+
Sbjct: 582 TRLQNV-TLGLTSSPWLSLPR 641
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.322 0.140 0.413
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,666,101
Number of Sequences: 28460
Number of extensions: 44057
Number of successful extensions: 216
Number of sequences better than 10.0: 26
Number of HSP's better than 10.0 without gapping: 214
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 215
length of query: 247
length of database: 4,897,600
effective HSP length: 88
effective length of query: 159
effective length of database: 2,393,120
effective search space: 380506080
effective search space used: 380506080
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 54 (25.4 bits)
Medicago: description of AC146554.3


