

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146527.7 - phase: 0
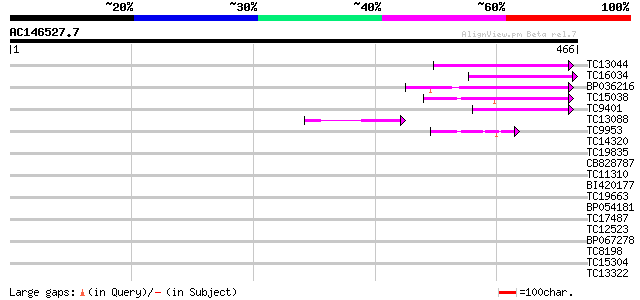
(466 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC13044 similar to UP|FKB7_WHEAT (Q43207) 70 kDa peptidylprolyl ... 78 3e-15
TC16034 homologue to UP|FKB2_VICFA (Q41649) FK506-binding protei... 76 1e-14
BP036216 74 7e-14
TC15038 similar to UP|O04287 (O04287) Immunophilin, complete 61 4e-10
TC9401 similar to UP|Q84LW7 (Q84LW7) Peptidylprolyl cis-trans is... 60 8e-10
TC13088 60 8e-10
TC9953 similar to GB|CAD35362.1|21535744|ATH490171 FK506 binding... 43 1e-04
TC14320 similar to UP|Q41111 (Q41111) Dehydrin, partial (56%) 37 0.005
TC19835 similar to UP|Q9ZRW0 (Q9ZRW0) Nucleolar protein (Fragmen... 37 0.009
CB828787 36 0.012
TC11310 similar to UP|BAD11331 (BAD11331) BRI1-KD interacting pr... 34 0.045
BI420177 34 0.045
TC19663 similar to UP|Q8H6S8 (Q8H6S8) Translation initiation fac... 34 0.045
BP054181 34 0.045
TC17487 UP|CAE45597 (CAE45597) SAR DNA-binding protein-like prot... 34 0.059
TC12523 33 0.077
BP067278 33 0.077
TC8198 similar to UP|P93273 (P93273) PAFD103 protein (Fragment),... 33 0.13
TC15304 32 0.22
TC13322 similar to UP|Q9ZS13 (Q9ZS13) RNA helicase (Fragment), p... 31 0.38
>TC13044 similar to UP|FKB7_WHEAT (Q43207) 70 kDa peptidylprolyl isomerase
(Peptidyl-prolyl cis-trans isomerase) (PPIase)
(Rotamase) , partial (26%)
Length = 531
Score = 78.2 bits (191), Expect = 3e-15
Identities = 37/115 (32%), Positives = 63/115 (54%)
Frame = +2
Query: 349 KPSQVRTLSNGLVIQELETGKENGKIAAIGKKISINYTGKLKENGVVVESNAGEAPFKFR 408
K + R + N + ++L + + +G ++ ++YTG L + S +APF F
Sbjct: 92 KVGEEREIGNSGLRKKLLKEGQGWETPEVGDEVQVHYTGTLLDGSKFDSSRDRDAPFSFT 271
Query: 409 LGKGEVIEGWDIGLEGMRVGEKRRLVVPPSMTSKKDGESENIPPNSWLVYDFELV 463
LG+G+VI+GWD G++ M+ GE +PP + + G IPPN+ L +D EL+
Sbjct: 272 LGQGQVIKGWDEGIKTMKKGENALFTIPPELAYGESGSPPTIPPNATLQFDVELL 436
>TC16034 homologue to UP|FKB2_VICFA (Q41649) FK506-binding protein 2
precursor (Peptidyl-prolyl cis-trans isomerase)
(PPIase) (Rotamase) (15 kDa FKBP) (FKBP-15) , partial
(91%)
Length = 592
Score = 75.9 bits (185), Expect = 1e-14
Identities = 38/89 (42%), Positives = 53/89 (58%)
Frame = +3
Query: 378 GKKISINYTGKLKENGVVVESNAGEAPFKFRLGKGEVIEGWDIGLEGMRVGEKRRLVVPP 437
G K+ ++Y GKL + V S +P F LG G+VI+GWD GL GM +GEKR+L +P
Sbjct: 168 GDKVKVHYRGKLTDGTVFDSSFERNSPIDFELGSGQVIKGWDQGLLGMCLGEKRKLKIPA 347
Query: 438 SMTSKKDGESENIPPNSWLVYDFELVKVH 466
+ + G IP + LV+D ELV V+
Sbjct: 348 KLGYGEQGSPPTIPGGATLVFDTELVGVN 434
>BP036216
Length = 574
Score = 73.6 bits (179), Expect = 7e-14
Identities = 42/142 (29%), Positives = 66/142 (45%), Gaps = 4/142 (2%)
Frame = +1
Query: 326 DVPVSAEQSSEMMKEDGNN----VEDTKPSQVRTLSNGLVIQELETGKENGKIAAIGKKI 381
D+P + + DG N V D K ++ L L+ + E G ++
Sbjct: 97 DIPAAGNDDFDFAGADGGNPMFKVGDEKEXGIQGLKKKLIKEG-----EGXDTPDAGDEV 261
Query: 382 SINYTGKLKENGVVVESNAGEAPFKFRLGKGEVIEGWDIGLEGMRVGEKRRLVVPPSMTS 441
++YTG L + S PF F LG+G+VI+GWD G++ M+ GE +P +
Sbjct: 262 HVHYTGTLLDGTKFDSSRDRGTPFNFTLGQGQVIKGWDEGIKTMKKGENALFTIPAELAY 441
Query: 442 KKDGESENIPPNSWLVYDFELV 463
+ G IPPN+ L +D EL+
Sbjct: 442 GESGSPPTIPPNATLQFDVELL 507
>TC15038 similar to UP|O04287 (O04287) Immunophilin, complete
Length = 602
Score = 60.8 bits (146), Expect = 4e-10
Identities = 40/128 (31%), Positives = 66/128 (51%), Gaps = 5/128 (3%)
Frame = +2
Query: 341 DGNNVEDTKPSQVRTLSNGLVIQELETGKENGKIAAIGKKISINYTGKLKENGVVVE--- 397
+G ++T+ S + S G+ Q L G G G+ ++++ TG K + +
Sbjct: 11 EGRETKETRSSSSSSSSMGVEKQLLRPG--TGPKPVPGQNVTVHCTGFGKNGDLSQQFWS 184
Query: 398 -SNAGEAPFKFRLGKGEVIEGWDIGLEGMRVGEKRRLVVPPSMTSKKDG-ESENIPPNSW 455
+ G+ PF F++G+G VI+GWD G+ GM+VGE RL P G + I PNS
Sbjct: 185 TKDPGQKPFTFKIGQGSVIKGWDEGVLGMQVGEVARLRCSPDYAYGAGGFPAWGIQPNSV 364
Query: 456 LVYDFELV 463
L ++ E++
Sbjct: 365 LDFEIEVL 388
>TC9401 similar to UP|Q84LW7 (Q84LW7) Peptidylprolyl cis-trans isomerase ,
partial (97%)
Length = 918
Score = 60.1 bits (144), Expect = 8e-10
Identities = 28/84 (33%), Positives = 48/84 (56%), Gaps = 1/84 (1%)
Frame = +1
Query: 381 ISINYTGKLKENGVVVES-NAGEAPFKFRLGKGEVIEGWDIGLEGMRVGEKRRLVVPPSM 439
+ ++Y G L + G V ++ + F F LGKG VI+ WD+ ++ M+VGE ++ P
Sbjct: 265 VDVHYEGTLADTGEVFDTTHEDNTIFSFELGKGSVIKAWDVAVKTMKVGEIAKITCKPEY 444
Query: 440 TSKKDGESENIPPNSWLVYDFELV 463
G +IPP++ LV++ EL+
Sbjct: 445 AYGSAGSPPDIPPDATLVFEVELL 516
>TC13088
Length = 401
Score = 60.1 bits (144), Expect = 8e-10
Identities = 38/84 (45%), Positives = 46/84 (54%), Gaps = 1/84 (1%)
Frame = +3
Query: 243 QDEKPKRKRKERSKEKTLFAADDACISNVVNLPQGNEHSNQDTVNGSVDNGAQDNPDGNQ 302
Q EKPK+KRK+R K +VD+GA+D PDGN
Sbjct: 57 QGEKPKKKRKDRPK--------------------------------TVDDGARDFPDGNL 140
Query: 303 SEDKKVKKKKKKSKSQGS-EVVNS 325
SED++VKK+KKKSKSQG EVVNS
Sbjct: 141 SEDREVKKRKKKSKSQGKVEVVNS 212
>TC9953 similar to GB|CAD35362.1|21535744|ATH490171 FK506 binding protein 1
{Arabidopsis thaliana;} , partial (29%)
Length = 481
Score = 43.1 bits (100), Expect = 1e-04
Identities = 28/75 (37%), Positives = 41/75 (54%), Gaps = 2/75 (2%)
Frame = +2
Query: 347 DTKPSQVRTLSNGLVIQELETGKENGKIAAIGKKISINYTGKLKENGVVVES--NAGEAP 404
+ P ++ +GL + G G A G+ I +Y G+L ENG + +S N G+ P
Sbjct: 266 EAPPCELTAAPSGLAFCDKLVG--TGPQAVKGQLIKAHYVGRL-ENGKIFDSSYNRGQ-P 433
Query: 405 FKFRLGKGEVIEGWD 419
FR+G GEVI+GWD
Sbjct: 434 LTFRVGVGEVIKGWD 478
>TC14320 similar to UP|Q41111 (Q41111) Dehydrin, partial (56%)
Length = 1231
Score = 37.4 bits (85), Expect = 0.005
Identities = 30/125 (24%), Positives = 56/125 (44%), Gaps = 3/125 (2%)
Frame = +3
Query: 211 LLTGNEQNQQQPDDKKAETTDKTLPSSQVGQGQDEKPKRKRKERSKEKTLFAADDACISN 270
++ QN+ + + ET D+ + + ++EKP+ + EK + +
Sbjct: 354 IMAEENQNKYESGTTEVETQDRGVFDFLGKKKEEEKPQEEVIVTEFEKVKVSEEPEKKKE 533
Query: 271 VVNLPQGNEHSN---QDTVNGSVDNGAQDNPDGNQSEDKKVKKKKKKSKSQGSEVVNSDV 327
V+ +HS + + S + + D +G + E+KK KKKKK+ K S N+ V
Sbjct: 534 EVDHEGEKKHSTLLEKLHRSDSSSSSSSDEEEGKEGEEKK-KKKKKEKKEVKSTDENTSV 710
Query: 328 PVSAE 332
PV +
Sbjct: 711 PVEVD 725
Score = 32.3 bits (72), Expect = 0.17
Identities = 29/106 (27%), Positives = 54/106 (50%), Gaps = 3/106 (2%)
Frame = +3
Query: 107 QEIVDNVDKEAVDDGKKDGEDRENITIETKLKTDNVVADIQTHREAEPSDQLAVPSTVPD 166
QE V + E V ++ + +E + E + K ++ + HR SD + S+ +
Sbjct: 465 QEEVIVTEFEKVKVSEEPEKKKEEVDHEGEKKHSTLLEKL--HR----SDSSSSSSSDEE 626
Query: 167 VGDMKKSKKKKKGKEKETKSSSNEHST---DLDNAAQDEPKMNMDE 209
G + KKKKK KEK+ S++E+++ ++D A +E K +D+
Sbjct: 627 EGKEGEEKKKKKKKEKKEVKSTDENTSVPVEVDPAHAEEKKGFLDK 764
Score = 28.5 bits (62), Expect = 2.5
Identities = 32/117 (27%), Positives = 49/117 (41%), Gaps = 3/117 (2%)
Frame = +3
Query: 14 KKDSEDHENSIIETKLKTDNAFTESQIHSREAGPSDHLVDPCTVPDVGDIKKSKKKKKGK 73
KK+ DHE K + E ++H ++ S + + G + KKKKK K
Sbjct: 525 KKEEVDHEGE------KKHSTLLE-KLHRSDSSSSSSSDE-----EEGKEGEEKKKKKKK 668
Query: 74 E-KETKSSSNGDS--TELDNAEQDEPKMNMAQDLLDQEIVDNVDKEAVDDGKKDGED 127
E KE KS+ S E+D A +E K +D + ++ GKK E+
Sbjct: 669 EKKEVKSTDENTSVPVEVDPAHAEEKK----------GFLDKIKEKLPGHGKKTTEE 809
>TC19835 similar to UP|Q9ZRW0 (Q9ZRW0) Nucleolar protein (Fragment), partial
(16%)
Length = 550
Score = 36.6 bits (83), Expect = 0.009
Identities = 28/126 (22%), Positives = 49/126 (38%), Gaps = 2/126 (1%)
Frame = +1
Query: 225 KKAETTDKTLPSSQVGQGQDEKPKRKRKERSKEKTLFAADDACISNVVNLPQGNEHSNQD 284
+ A D + + +V K +K+++ + E A D ++N + +
Sbjct: 46 ESAANNDAEMETEEVPVEASSKKSKKKQKAADEGDEMAVDKPTVTNGDAVEDHKSEKKKK 225
Query: 285 TVNGSVDN--GAQDNPDGNQSEDKKVKKKKKKSKSQGSEVVNSDVPVSAEQSSEMMKEDG 342
+D A++ +GN SE + VKKKK + G V A S+ K+
Sbjct: 226 KEKRKLDQEVAAENGTNGNASEPRSVKKKKDRKDDNGE-------AVEAASESKKKKKSK 384
Query: 343 NNVEDT 348
N E T
Sbjct: 385 KNAE*T 402
Score = 35.0 bits (79), Expect = 0.026
Identities = 25/111 (22%), Positives = 52/111 (46%), Gaps = 10/111 (9%)
Frame = +1
Query: 164 VPDVGDMKKSKKKKKGKEKETKSSSNEHSTDLDNAAQD--------EPKMNMDEDLL--T 213
VP KKSKKK+K ++ + + ++ + +A +D + K +D+++
Sbjct: 88 VPVEASSKKSKKKQKAADEGDEMAVDKPTVTNGDAVEDHKSEKKKKKEKRKLDQEVAAEN 267
Query: 214 GNEQNQQQPDDKKAETTDKTLPSSQVGQGQDEKPKRKRKERSKEKTLFAAD 264
G N +P K + D+ + + + E K+K+ +++ E T+ AD
Sbjct: 268 GTNGNASEPRSVK-KKKDRKDDNGEAVEAASESKKKKKSKKNAE*TMLVAD 417
Score = 28.9 bits (63), Expect = 1.9
Identities = 27/102 (26%), Positives = 43/102 (41%), Gaps = 12/102 (11%)
Frame = +1
Query: 45 AGPSDHLVDPCTVPDVGDIKKSKKKKKGKEK------ETKSSSNGDSTELDNAEQDEPKM 98
A +D ++ VP KKSKKK+K ++ + + +NGD+ E + E K
Sbjct: 52 AANNDAEMETEEVPVEASSKKSKKKQKAADEGDEMAVDKPTVTNGDAVE---DHKSEKKK 222
Query: 99 NMAQDLLDQEIV------DNVDKEAVDDGKKDGEDRENITIE 134
+ LDQE+ N + KKD +D +E
Sbjct: 223 KKEKRKLDQEVAAENGTNGNASEPRSVKKKKDRKDDNGEAVE 348
>CB828787
Length = 343
Score = 36.2 bits (82), Expect = 0.012
Identities = 20/44 (45%), Positives = 28/44 (63%)
Frame = +1
Query: 422 LEGMRVGEKRRLVVPPSMTSKKDGESENIPPNSWLVYDFELVKV 465
+EGMRVG KR ++VPP K G +E IPP + + EL++V
Sbjct: 205 VEGMRVGGKRTVLVPPENGYGKRGMNE-IPPGATFEMNIELLQV 333
>TC11310 similar to UP|BAD11331 (BAD11331) BRI1-KD interacting protein 102
(Fragment), partial (13%)
Length = 667
Score = 34.3 bits (77), Expect = 0.045
Identities = 24/96 (25%), Positives = 46/96 (47%)
Frame = +1
Query: 244 DEKPKRKRKERSKEKTLFAADDACISNVVNLPQGNEHSNQDTVNGSVDNGAQDNPDGNQS 303
++K K+K+K++ +E A+ D + E + V++G+ PD +QS
Sbjct: 79 EKKEKKKKKKKDQENGEVASSDE---------EKAEKEKKKKHKVKVEDGS---PDLDQS 222
Query: 304 EDKKVKKKKKKSKSQGSEVVNSDVPVSAEQSSEMMK 339
E KK KKK + ++ +E+ N +E+ + K
Sbjct: 223 EKKKKKKKDQDAEDNAAEISNGKEDRKSEKKHKKKK 330
Score = 32.3 bits (72), Expect = 0.17
Identities = 26/103 (25%), Positives = 47/103 (45%), Gaps = 18/103 (17%)
Frame = +1
Query: 118 VDDGKK----DGEDRENITIETKLKTDNVVA---DIQTHREAEPSDQLAVPSTVPDVGDM 170
VD+GK +GE +E + K + + VA + + +E + ++ V PD+
Sbjct: 43 VDEGKVKVEVEGEKKEKKKKKKKDQENGEVASSDEEKAEKEKKKKHKVKVEDGSPDLDQS 222
Query: 171 KKSKKKKKGKE-----------KETKSSSNEHSTDLDNAAQDE 202
+K KKKKK ++ KE + S +H + A++E
Sbjct: 223 EKKKKKKKDQDAEDNAAEISNGKEDRKSEKKHKKKKNQDAEEE 351
Score = 29.3 bits (64), Expect = 1.4
Identities = 21/78 (26%), Positives = 38/78 (47%), Gaps = 3/78 (3%)
Frame = +1
Query: 64 KKSKKKKKGKEKETKSSSNGDSTELDNAEQDEPKMNMAQDLLDQ---EIVDNVDKEAVDD 120
+K KKKKK +E +SS+ + E + ++ + K+ LDQ + D++A D+
Sbjct: 88 EKKKKKKKDQENGEVASSDEEKAEKEKKKKHKVKVEDGSPDLDQSEKKKKKKKDQDAEDN 267
Query: 121 GKKDGEDRENITIETKLK 138
+ +E+ E K K
Sbjct: 268 AAEISNGKEDRKSEKKHK 321
Score = 28.9 bits (63), Expect = 1.9
Identities = 19/49 (38%), Positives = 25/49 (50%)
Frame = +1
Query: 302 QSEDKKVKKKKKKSKSQGSEVVNSDVPVSAEQSSEMMKEDGNNVEDTKP 350
+ E K+ KKKKKK + G EV +SD + E K+ VED P
Sbjct: 73 EGEKKEKKKKKKKDQENG-EVASSD---EEKAEKEKKKKHKVKVEDGSP 207
Score = 26.9 bits (58), Expect = 7.2
Identities = 17/60 (28%), Positives = 30/60 (49%), Gaps = 2/60 (3%)
Frame = +1
Query: 58 PDVGDIKKSKKKKKGKEKETKSS--SNGDSTELDNAEQDEPKMNMAQDLLDQEIVDNVDK 115
PD+ +K KKKKK ++ E ++ SNG + + K A++ ++ I+ DK
Sbjct: 205 PDLDQSEKKKKKKKDQDAEDNAAEISNGKEDRKSEKKHKKKKNQDAEEE*NRGIILCFDK 384
>BI420177
Length = 444
Score = 34.3 bits (77), Expect = 0.045
Identities = 25/87 (28%), Positives = 38/87 (42%), Gaps = 6/87 (6%)
Frame = +3
Query: 125 GEDRENITIETKLKTDNVVADIQTH------REAEPSDQLAVPSTVPDVGDMKKSKKKKK 178
G D E I I+ + +D QT E +PSD L+ D KKS K++
Sbjct: 147 GADTEPIPIDVPPVVTSTPSDTQTEPQNDGRSEPDPSDDLSSEEAASD--GSKKSPKREL 320
Query: 179 GKEKETKSSSNEHSTDLDNAAQDEPKM 205
+E++ S + + L A QD P +
Sbjct: 321 DEEQQPASKKQKPLSSLSAANQDSPAL 401
Score = 26.9 bits (58), Expect = 7.2
Identities = 21/64 (32%), Positives = 28/64 (42%), Gaps = 1/64 (1%)
Frame = +3
Query: 36 TESQIHSR-EAGPSDHLVDPCTVPDVGDIKKSKKKKKGKEKETKSSSNGDSTELDNAEQD 94
TE Q R E PSD L D KKS K++ +E++ S + L A QD
Sbjct: 216 TEPQNDGRSEPDPSDDLSSEEAASDGS--KKSPKRELDEEQQPASKKQKPLSSLSAANQD 389
Query: 95 EPKM 98
P +
Sbjct: 390 SPAL 401
>TC19663 similar to UP|Q8H6S8 (Q8H6S8) Translation initiation factor,
partial (10%)
Length = 485
Score = 34.3 bits (77), Expect = 0.045
Identities = 29/114 (25%), Positives = 47/114 (40%), Gaps = 25/114 (21%)
Frame = +3
Query: 172 KSKKKKKGKEKETK-----------------------SSSNEHSTDLDNAAQDEPKMNMD 208
+ KKKKK KEKE K S NE T + + M
Sbjct: 36 QKKKKKKEKEKEKKAAAAAAGSAPANETVEVKAEVVESKKNESKTKAADKKLPKHVREMQ 215
Query: 209 EDLLTGNE--QNQQQPDDKKAETTDKTLPSSQVGQGQDEKPKRKRKERSKEKTL 260
E L E + +++ +++K ++ + + Q E+ +R++KER KEK L
Sbjct: 216 EALARRKEAEERKKREEEEKLRKEEEERRRQEELERQAEEARRRKKEREKEKLL 377
Score = 33.9 bits (76), Expect = 0.059
Identities = 32/150 (21%), Positives = 67/150 (44%), Gaps = 9/150 (6%)
Frame = +3
Query: 64 KKSKKKKKGKEKETKSSSNGDS---------TELDNAEQDEPKMNMAQDLLDQEIVDNVD 114
KK KKK+K KEK+ +++ G + E+ ++++E K A L + + + +
Sbjct: 39 KKKKKKEKEKEKKAAAAAAGSAPANETVEVKAEVVESKKNESKTKAADKKLPKHVREMQE 218
Query: 115 KEAVDDGKKDGEDRENITIETKLKTDNVVADIQTHREAEPSDQLAVPSTVPDVGDMKKSK 174
A +K+ E+R+ E KL+ + + R E ++ A ++++
Sbjct: 219 ALA---RRKEAEERKKREEEEKLRKEE-----EERRRQEELERQA-----------EEAR 341
Query: 175 KKKKGKEKETKSSSNEHSTDLDNAAQDEPK 204
++KK +EKE + L ++E +
Sbjct: 342 RRKKEREKEKLLKKKQEGKLLTGKQKEEQR 431
>BP054181
Length = 471
Score = 34.3 bits (77), Expect = 0.045
Identities = 23/78 (29%), Positives = 39/78 (49%)
Frame = -1
Query: 244 DEKPKRKRKERSKEKTLFAADDACISNVVNLPQGNEHSNQDTVNGSVDNGAQDNPDGNQS 303
+ K KRK+K++ + +D +S+ +LP N HS N ++D+ P N+
Sbjct: 435 ETKGKRKKKQKERAIDDDVVEDLVLSSDDDLPSSNSHSAGK--NDAIDHLP---PKQNKK 271
Query: 304 EDKKVKKKKKKSKSQGSE 321
+ +K K K K+ KS E
Sbjct: 270 QKRKQKHKTKRLKSHVEE 217
>TC17487 UP|CAE45597 (CAE45597) SAR DNA-binding protein-like protein, complete
Length = 1794
Score = 33.9 bits (76), Expect = 0.059
Identities = 36/142 (25%), Positives = 63/142 (44%)
Frame = +1
Query: 116 EAVDDGKKDGEDRENITIETKLKTDNVVADIQTHREAEPSDQLAVPSTVPDVGDMKKSKK 175
EA D +K G + T KT N AD ++++ S + + + K
Sbjct: 1381 EAYDKDRKKGAGG----LITPAKTYNPSADAVISQKSD--------SAMDEDTHEPSADK 1524
Query: 176 KKKGKEKETKSSSNEHSTDLDNAAQDEPKMNMDEDLLTGNEQNQQQPDDKKAETTDKTLP 235
KK+ KEK+ K NE +D P + DED G+E+ + +KK + + T
Sbjct: 1525 KKEKKEKKKKKEKNEE--------KDVPLL-ADED---GDEEKEVVKKEKKKKRKEST-E 1665
Query: 236 SSQVGQGQDEKPKRKRKERSKE 257
+ ++ G D K+K++++ E
Sbjct: 1666 NVELQNGDDAGEKKKKRKKHAE 1731
>TC12523
Length = 603
Score = 33.5 bits (75), Expect = 0.077
Identities = 28/100 (28%), Positives = 49/100 (49%), Gaps = 1/100 (1%)
Frame = +2
Query: 227 AETTDKTLPSSQVGQGQDEKPKRKRKERSKEKTLFA-ADDACISNVVNLPQGNEHSNQDT 285
AE DK L S+ G D K K K +++ E +++ CI + ++H + D
Sbjct: 281 AEKKDKKLEKSK---GGDTKNKEKENQKNSEPVGDGGSNEDCIDAA---GEDSDHCDDDV 442
Query: 286 VNGSVDNGAQDNPDGNQSEDKKVKKKKKKSKSQGSEVVNS 325
+ +G+ +G + + KK+KKKK K ++ GS N+
Sbjct: 443 CGDA--SGSGGGGEGGKKK-KKIKKKKNKGENGGSAPPNN 553
>BP067278
Length = 562
Score = 33.5 bits (75), Expect = 0.077
Identities = 27/122 (22%), Positives = 49/122 (40%)
Frame = +1
Query: 138 KTDNVVADIQTHREAEPSDQLAVPSTVPDVGDMKKSKKKKKGKEKETKSSSNEHSTDLDN 197
K + ++ + HRE + S ++V D K ++ + K +S S
Sbjct: 13 KDEEILQEKSKHRERKRSRSVSV--------DEKPQRRSRSSPRKVDESGSRHKKRSRSK 168
Query: 198 AAQDEPKMNMDEDLLTGNEQNQQQPDDKKAETTDKTLPSSQVGQGQDEKPKRKRKERSKE 257
+ +D K N E L N + + +DK+ + T Q +DE+ K K R +
Sbjct: 169 SVED--KHNFPEKL-EENRIRKSRHNDKRHSRSRSTESRDQTDAREDERKYGKSKHRDSK 339
Query: 258 KT 259
+T
Sbjct: 340 RT 345
>TC8198 similar to UP|P93273 (P93273) PAFD103 protein (Fragment), partial
(65%)
Length = 850
Score = 32.7 bits (73), Expect = 0.13
Identities = 36/163 (22%), Positives = 70/163 (42%), Gaps = 22/163 (13%)
Frame = +3
Query: 245 EKPKRKRKERSKEKTLFAADDACISNVVNLPQGNEHSNQDTVNGSVDN-GAQDNPDGNQS 303
++ +R+ + ++K A +A +++ P+ + +T D G + + DG +
Sbjct: 51 KETERQLSXKERKKKELAELEALLADFGVAPKESTDGQDETQGAPQDKKGDEADGDGEKK 230
Query: 304 E------DKKVKKKKKKSKSQGSEVVNSDVPVSAEQS-----SEMMKEDGNNVE-DTKPS 351
E K KKKKKK K+ + D P S++ + +E +ED + V+ +
Sbjct: 231 ETTAAGGSKNAKKKKKKDKASKEVKESQDQPNSSDTNNGPDLAENPEEDASTVDVKERLK 410
Query: 352 QVRTLSNGLVIQEL---------ETGKENGKIAAIGKKISINY 385
+V +L +E+ E + K+AA KK +Y
Sbjct: 411 KVASLKKKKSSKEMDAAAKAAAQEAAARHAKLAAAKKKEKNHY 539
Score = 26.6 bits (57), Expect = 9.4
Identities = 22/93 (23%), Positives = 43/93 (45%), Gaps = 2/93 (2%)
Frame = +3
Query: 168 GDMKKSKKKKKGK-EKETKSSSNEHSTDLDNAAQDEPKMNMDEDLLTGN-EQNQQQPDDK 225
G KKKKK K KE K S ++ ++ N D + N +ED T + ++ ++
Sbjct: 249 GSKNAKKKKKKDKASKEVKESQDQPNSSDTNNGPDLAE-NPEEDASTVDVKERLKKVASL 425
Query: 226 KAETTDKTLPSSQVGQGQDEKPKRKRKERSKEK 258
K + + K + ++ Q+ + + +K+K
Sbjct: 426 KKKKSSKEMDAAAKAAAQEAAARHAKLAAAKKK 524
>TC15304
Length = 477
Score = 32.0 bits (71), Expect = 0.22
Identities = 20/57 (35%), Positives = 28/57 (49%)
Frame = +1
Query: 174 KKKKKGKEKETKSSSNEHSTDLDNAAQDEPKMNMDEDLLTGNEQNQQQPDDKKAETT 230
KKKK K+++TK SSN ++ ++AQ EP+M L DD TT
Sbjct: 289 KKKKSDKDRKTKGSSNSKTS--SSSAQSEPQMFWAPAPLNATSWADVDDDDDYYATT 453
Score = 30.4 bits (67), Expect = 0.65
Identities = 18/50 (36%), Positives = 29/50 (58%)
Frame = +1
Query: 67 KKKKKGKEKETKSSSNGDSTELDNAEQDEPKMNMAQDLLDQEIVDNVDKE 116
KKKK K+++TK SSN ++ ++ Q EP+M A L+ +VD +
Sbjct: 289 KKKKSDKDRKTKGSSNSKTS--SSSAQSEPQMFWAPAPLNATSWADVDDD 432
>TC13322 similar to UP|Q9ZS13 (Q9ZS13) RNA helicase (Fragment), partial (7%)
Length = 528
Score = 31.2 bits (69), Expect = 0.38
Identities = 23/86 (26%), Positives = 41/86 (46%), Gaps = 5/86 (5%)
Frame = +3
Query: 278 NEHSNQDTVNGSVDNGAQDNPDGNQSEDKKVKKKKK---KSKSQGSEVVNSDVPVSAEQS 334
++H N + + DN +G +SE KK KKK+K KS+ E ++ +A+++
Sbjct: 159 DKHQNGEKTSPKRKLEELDNQNGTESEKKKKKKKEKHDNKSEENAEETKEANGNGNADET 338
Query: 335 SEMMKE--DGNNVEDTKPSQVRTLSN 358
E G + D K + V+T +
Sbjct: 339 VADGAEVVTGKDAADAKYAAVKTFKD 416
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.302 0.124 0.332
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,672,526
Number of Sequences: 28460
Number of extensions: 60892
Number of successful extensions: 672
Number of sequences better than 10.0: 80
Number of HSP's better than 10.0 without gapping: 451
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 603
length of query: 466
length of database: 4,897,600
effective HSP length: 93
effective length of query: 373
effective length of database: 2,250,820
effective search space: 839555860
effective search space used: 839555860
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 17 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 43 (21.7 bits)
S2: 57 (26.6 bits)
Medicago: description of AC146527.7


