

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146343.9 + phase: 0
(436 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
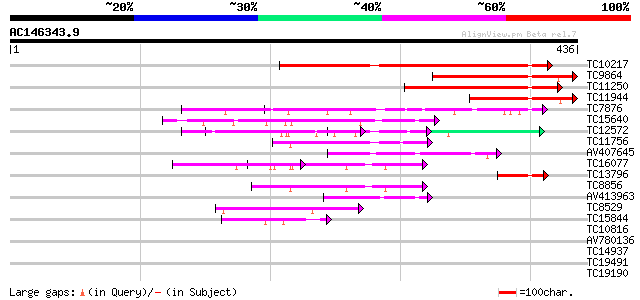
Score E
Sequences producing significant alignments: (bits) Value
TC10217 homologue to UP|AAQ54906 (AAQ54906) Cell cycle switch pr... 209 9e-55
TC9864 weakly similar to UP|Q24044 (Q24044) Fizzy protein, parti... 173 6e-44
TC11250 homologue to UP|AAQ72359 (AAQ72359) B-type cell cycle sw... 142 8e-35
TC11944 weakly similar to UP|Q9W4H9 (Q9W4H9) FZR protein (GH0762... 129 1e-30
TC7876 homologue to UP|GBLP_SOYBN (Q39836) Guanine nucleotide-bi... 56 1e-08
TC15640 similar to GB|AAP37794.1|30725544|BT008435 At3g63460 {Ar... 52 1e-07
TC12572 similar to PIR|T46032|T46032 WD-40 repeat regulatory pro... 51 4e-07
TC11756 weakly similar to GB|AAH01494.1|16306637|BC001494 U5 snR... 48 3e-06
AV407645 47 5e-06
TC16077 similar to PIR|T00593|T02480 sec13-related protein At2g3... 46 1e-05
TC13796 homologue to UP|Q9M7I2 (Q9M7I2) WD-repeat cell cycle reg... 45 2e-05
TC8856 similar to UP|Q8W403 (Q8W403) Sec13p, partial (48%) 45 3e-05
AV413963 44 5e-05
TC8529 homologue to UP|Q94KS2 (Q94KS2) TGF-beta receptor-interac... 43 9e-05
TC15844 homologue to UP|Q9FV61 (Q9FV61) Heterotrimeric GTP-bindi... 40 6e-04
TC10816 homologue to UP|CAE45585 (CAE45585) Coatomer alpha subun... 39 0.001
AV780136 39 0.002
TC14937 similar to UP|Q9FN19 (Q9FN19) Genomic DNA, chromosome 5,... 38 0.004
TC19491 similar to GB|AAP37692.1|30725340|BT008333 At4g29830 {Ar... 37 0.006
TC19190 weakly similar to PIR|T46032|T46032 WD-40 repeat regulat... 37 0.006
>TC10217 homologue to UP|AAQ54906 (AAQ54906) Cell cycle switch protein
CCS52a, partial (45%)
Length = 640
Score = 209 bits (531), Expect = 9e-55
Identities = 99/210 (47%), Positives = 138/210 (65%)
Frame = +2
Query: 208 VGSLAWNGHVLTTGGMDGKIVNNDVRLRSQIINTYRGHRREVCGLKWSLDGKQLASGGND 267
VG+LA +L++GG D I +R + ++ GH+ VCGLKWS D ++LASGGND
Sbjct: 2 VGALAXXSSLLSSGGRDKNIYQRXIRAQEXYVSKLSGHKSXVCGLKWSYDNRELASGGND 181
Query: 268 NVVHIWDMSAVSSNSPTRWLYRFDEHKAAVKALAWCPFQGNLLASGGGGGDCCVKLWNTG 327
N + +W+ + T+ + ++ EH AAVKA+AW P LLASGGG D C++ WNT
Sbjct: 182 NRLFVWNQHS------TQPVLKYCEHTAAVKAIAWSPHLHGLLASGGGTADRCIRFWNTT 343
Query: 328 MGERMNSVDTGSQVCALLWSKNERELLSSHGLTQNQLTLWKYPSMLKIAELHGHTSRVLH 387
++ +DTGSQVC L+WSKN EL+S+HG +QNQ+ +W+YP+M K+A L GHT RVL+
Sbjct: 344 TNSHLSCMDTGSQVCNLVWSKNVNELVSTHGYSQNQIIVWRYPTMSKLATLTGHTYRVLY 523
Query: 388 MTQSPDGSTVASAAAAADQTLRFWEVFGTP 417
+ SPDG T+ + A D+TL FW VF P
Sbjct: 524 LAISPDGQTIVT--GAGDETLXFWNVFPFP 607
>TC9864 weakly similar to UP|Q24044 (Q24044) Fizzy protein, partial (14%)
Length = 657
Score = 173 bits (438), Expect = 6e-44
Identities = 87/113 (76%), Positives = 97/113 (84%), Gaps = 2/113 (1%)
Frame = +2
Query: 326 TGMGERMNSVDTGSQVCALLWSKNERELLSSHGLTQNQLTLWKYPSMLKIAELHGHTSRV 385
T G +N+++TGSQVCALLWSK+ERELLSSHG TQNQLTLWKYPSMLK+AEL+GHTSRV
Sbjct: 2 THTGACLNTINTGSQVCALLWSKSERELLSSHGFTQNQLTLWKYPSMLKMAELNGHTSRV 181
Query: 386 LHMTQSPDGSTVASAAAAADQTLRFWEVFGTPPAA--PPKRNKMPFADSNRIR 436
LHM QSPDG TVAS AAAD+TLRFW VFGTP A+ PKRN+ PFAD NRIR
Sbjct: 182 LHMAQSPDGCTVAS--AAADETLRFWNVFGTPEASKPSPKRNREPFADFNRIR 334
>TC11250 homologue to UP|AAQ72359 (AAQ72359) B-type cell cycle switch
protein ccs52B, partial (28%)
Length = 564
Score = 142 bits (359), Expect = 8e-35
Identities = 64/122 (52%), Positives = 85/122 (69%)
Frame = +2
Query: 304 PFQGNLLASGGGGGDCCVKLWNTGMGERMNSVDTGSQVCALLWSKNERELLSSHGLTQNQ 363
P Q NLL SGGG D C++ WNT G ++N VDTGSQVC L WSKN E++S+HG +QNQ
Sbjct: 41 PHQSNLLVSGGGTADRCIRFWNTTNGHQLNHVDTGSQVCNLAWSKNVNEIVSTHGYSQNQ 220
Query: 364 LTLWKYPSMLKIAELHGHTSRVLHMTQSPDGSTVASAAAAADQTLRFWEVFGTPPAAPPK 423
+ +WKYPS+ K+A L GH+ RVL++ SPDG T+ + A D+TLRFW VF + P
Sbjct: 221 IMVWKYPSLSKVATLTGHSMRVLYLAMSPDGQTIVT--GAGDETLRFWNVFPSMKTPAPV 394
Query: 424 RN 425
++
Sbjct: 395 KD 400
Score = 37.0 bits (84), Expect = 0.006
Identities = 28/113 (24%), Positives = 50/113 (43%), Gaps = 6/113 (5%)
Frame = +2
Query: 169 CWAPDGRHLAV---GLTNSHVQLWDTAANKQLRTLKGGHRARVGSLAWNGHVLTTGGMDG 225
C +P +L V G + ++ W+T QL + G ++V +LAW+ +V G
Sbjct: 32 CLSPHQSNLLVSGGGTADRCIRFWNTTNGHQLNHVDTG--SQVCNLAWSKNVNEIVSTHG 205
Query: 226 KIVNNDVRLRSQIIN---TYRGHRREVCGLKWSLDGKQLASGGNDNVVHIWDM 275
N + + ++ T GH V L S DG+ + +G D + W++
Sbjct: 206 YSQNQIMVWKYPSLSKVATLTGHSMRVLYLAMSPDGQTIVTGAGDETLRFWNV 364
>TC11944 weakly similar to UP|Q9W4H9 (Q9W4H9) FZR protein (GH07620p)
(RE20929p), partial (12%)
Length = 512
Score = 129 bits (323), Expect = 1e-30
Identities = 66/85 (77%), Positives = 72/85 (84%), Gaps = 2/85 (2%)
Frame = +1
Query: 354 LSSHGLTQNQLTLWKYPSMLKIAELHGHTSRVLHMTQSPDGSTVASAAAAADQTLRFWEV 413
LSSHG TQNQLTLWKYPSMLK+AEL+GHTSRVL+M QSPDG TVAS AAAD+TLRFW V
Sbjct: 1 LSSHGFTQNQLTLWKYPSMLKMAELNGHTSRVLYMAQSPDGCTVAS--AAADETLRFWNV 174
Query: 414 FGTPPAAPP--KRNKMPFADSNRIR 436
FGTP A+ P K N+ PFAD NRIR
Sbjct: 175 FGTPEASKPSSKPNREPFADFNRIR 249
>TC7876 homologue to UP|GBLP_SOYBN (Q39836) Guanine nucleotide-binding
protein beta subunit-like protein, complete
Length = 1379
Score = 56.2 bits (134), Expect = 1e-08
Identities = 74/306 (24%), Positives = 127/306 (41%), Gaps = 25/306 (8%)
Frame = +3
Query: 133 VLSIALDHTIYFWNASDSSGSEFVTVDEEEGP---VTSVCWAPDGRHLAVGLTNSHVQLW 189
+++ + D +I W+ + + V G V V + DG+ G + ++LW
Sbjct: 162 IVTASRDKSIILWHLTKEDKTYGVPRRRLTGHSHFVQDVVLSSDGQFALSGSWDGELRLW 341
Query: 190 DTAANKQLRTLKGGHRARVGSLAW--NGHVLTTGGMDGKI-VNNDVRLRSQIINTYRGHR 246
D AA R GH V S+A+ + + + D I + N + I H
Sbjct: 342 DLAAGTSARRFV-GHTKDVLSVAFSIDNRQIVSASRDRTIKLWNTLGECKYTIQDNDAHS 518
Query: 247 REVCGLKWSLDGKQ--LASGGNDNVVHIWDMSAVSSNSPTRWLYRFDEHKAAVKALAWCP 304
V +++S Q + S D V +W+++ + H V +A P
Sbjct: 519 DWVSCVRFSPSTLQPTIVSASWDRTVKVWNLTNCKLRN------TLAGHSGYVNTVAVSP 680
Query: 305 FQGNLLASGGGGGDCCVKLWNTGMGERMNSVDTGSQVCALLWSKNERELLSSHGLTQNQL 364
G+L ASGG G + LW+ G+R+ S+D GS + AL +S N L ++ T+ +
Sbjct: 681 -DGSLCASGGKDG--VILLWDLAEGKRLYSLDAGSIIHALCFSPNRYWLCAA---TETSI 842
Query: 365 TLWKYPSMLKIAELH------------GHTS--RVLHMTQ---SPDGSTVASAAAAADQT 407
+W S + +L G+T+ +V++ T S DGST+ S D
Sbjct: 843 KIWDLESKSIVEDLKVDLKTEADATTGGNTNKKKVIYCTSLNWSADGSTLFS--GYTDGV 1016
Query: 408 LRFWEV 413
+R W +
Sbjct: 1017IRVWAI 1034
Score = 41.6 bits (96), Expect = 3e-04
Identities = 52/228 (22%), Positives = 90/228 (38%), Gaps = 11/228 (4%)
Frame = +3
Query: 197 LRTLKGGHRARVGSLAW---NGHVLTTGGMDGKIVNNDVRLRSQIINTYR----GHRREV 249
LR H +V ++A N ++ T D I+ + + R GH V
Sbjct: 90 LRGTMRAHTDQVTAIATPIDNSDMIVTASRDKSIILWHLTKEDKTYGVPRRRLTGHSHFV 269
Query: 250 CGLKWSLDGKQLASGGNDNVVHIWDMSAVSSNSPTRWLYRFDEHKAAVKALAWCPFQGNL 309
+ S DG+ SG D + +WD++A +S RF H V ++A+ +
Sbjct: 270 QDVVLSSDGQFALSGSWDGELRLWDLAAGTS------ARRFVGHTKDVLSVAFSIDNRQI 431
Query: 310 LASGGGGGDCCVKLWNTGMGERMNSVDTGSQ----VCALLWSKNERELLSSHGLTQNQLT 365
+++ D +KLWNT +GE ++ V + +S + + +
Sbjct: 432 VSA---SRDRTIKLWNT-LGECKYTIQDNDAHSDWVSCVRFSPSTLQPTIVSASWDRTVK 599
Query: 366 LWKYPSMLKIAELHGHTSRVLHMTQSPDGSTVASAAAAADQTLRFWEV 413
+W + L GH+ V + SPDGS AS D + W++
Sbjct: 600 VWNLTNCKLRNTLAGHSGYVNTVAVSPDGSLCAS--GGKDGVILLWDL 737
>TC15640 similar to GB|AAP37794.1|30725544|BT008435 At3g63460 {Arabidopsis
thaliana;}, partial (26%)
Length = 1022
Score = 52.4 bits (124), Expect = 1e-07
Identities = 61/234 (26%), Positives = 99/234 (42%), Gaps = 21/234 (8%)
Frame = +2
Query: 118 SDDFSLNLLDWGSRNVLSIALDHTIYFWNA-----SDSSGSEFVTVDEEEGPVTSVCW-- 170
SD FSL L+ G +D I WN S++ + + +GPV + +
Sbjct: 368 SDGFSLGLVAGG-------LVDGNIDIWNPLSLIRSENESALVGHLVRHKGPVRGLEFNT 526
Query: 171 -APDGRHLAVGLTNSHVQLWDTAANKQ---LRTLKGGHRARVGS---LAWNG---HVLTT 220
AP+ LA G + + +WD A + LKG A G L+WN H+L +
Sbjct: 527 IAPN--LLASGAEDGEICIWDLANPSEPTHFPPLKGSGSASQGEISFLSWNSKVQHILAS 700
Query: 221 GGMDGKIVNNDVRLRSQIINTYRGHRREVCGLKWSLD-GKQLASGGNDN---VVHIWDMS 276
+G V D++ + +I+ RR L+W+ D QL +++ + +WDM
Sbjct: 701 TSYNGTTVVWDLKKQKPVISFADSVRRRCSVLQWNPDIATQLVVASDEDGSPSLRLWDMR 880
Query: 277 AVSSNSPTRWLYRFDEHKAAVKALAWCPFQGNLLASGGGGGDCCVKLWNTGMGE 330
+ + + F H V A++WCP + L + G D W+T GE
Sbjct: 881 NIITP-----IKEFVGHTRGVIAMSWCPNDSSYLLT--CGKDSRTICWDTISGE 1021
Score = 35.8 bits (81), Expect = 0.014
Identities = 37/165 (22%), Positives = 66/165 (39%), Gaps = 12/165 (7%)
Frame = +2
Query: 261 LASGGNDNVVHIWD-MSAVSSNSPTRWLYRFDEHKAAVKALAWCPFQGNLLASGGGGGDC 319
+A G D + IW+ +S + S + + + HK V+ L + NLLASG G+
Sbjct: 392 VAGGLVDGNIDIWNPLSLIRSENESALVGHLVRHKGPVRGLEFNTIAPNLLASGAEDGEI 571
Query: 320 CVKLWNTGMGER------MNSVDTGSQ--VCALLWSKNERELLSSHGLTQNQLTLWKYPS 371
C+ W+ + + SQ + L W+ + +L+S +W
Sbjct: 572 CI--WDLANPSEPTHFPPLKGSGSASQGEISFLSWNSKVQHILASTSY-NGTTVVWDLKK 742
Query: 372 MLKIAELHGHTSRVLHMTQ-SPDGSTVASAAAAAD--QTLRFWEV 413
+ R + Q +PD +T A+ D +LR W++
Sbjct: 743 QKPVISFADSVRRRCSVLQWNPDIATQLVVASDEDGSPSLRLWDM 877
>TC12572 similar to PIR|T46032|T46032 WD-40 repeat regulatory protein tup1
homolog - Arabidopsis thaliana
{Arabidopsis thaliana;}, partial (51%)
Length = 770
Score = 50.8 bits (120), Expect = 4e-07
Identities = 38/147 (25%), Positives = 72/147 (48%), Gaps = 6/147 (4%)
Frame = +3
Query: 133 VLSIALDHTIYFWNASDSSGSEFVTVDEEEGPVTSVCWAPDGRHLAVGLTNSHVQLWDTA 192
++S + D W+AS + + +D+E PV+ V ++P+ + + VG +++++LW+ +
Sbjct: 84 IVSSSYDGLCRIWDASTGHCMKTL-IDDENPPVSFVKFSPNAKFILVGTLDNNLRLWNYS 260
Query: 193 ANKQLRTLKGGHRAR--VGSL--AWNGHVLTTGGMDGKIVNNDVRLRSQIINTYRGHRRE 248
+ L+T G ++ + S NG + G D I +++ R +I+ GH
Sbjct: 261 TGRFLKTYTGHVNSKYCISSTFSTTNGKYVVGGSEDHGIYLWELQTR-KIVQKLEGHSDT 437
Query: 249 VCGLKWSLDGKQLASG--GNDNVVHIW 273
V + +ASG GND V IW
Sbjct: 438 VVSVSCHPTENMIASGALGNDKTVKIW 518
Score = 49.3 bits (116), Expect = 1e-06
Identities = 40/183 (21%), Positives = 79/183 (42%), Gaps = 9/183 (4%)
Frame = +3
Query: 151 SGSEFVTVDEEEGPVTSVCWAPDGRHLAVGLTNSHVQLWDTAANKQLRTLKGGHRARVGS 210
SG + PVT+V + DG + + ++WD + ++TL V
Sbjct: 6 SGKCLKVLPAHSDPVTAVDFNRDGSLIVSSSYDGLCRIWDASTGHCMKTLIDDENPPVSF 185
Query: 211 LAW--NGHVLTTGGMDGKIVNNDVRL----RSQIINTYRGHRRE---VCGLKWSLDGKQL 261
+ + N + G +D N++RL + + TY GH + + +GK +
Sbjct: 186 VKFSPNAKFILVGTLD-----NNLRLWNYSTGRFLKTYTGHVNSKYCISSTFSTTNGKYV 350
Query: 262 ASGGNDNVVHIWDMSAVSSNSPTRWLYRFDEHKAAVKALAWCPFQGNLLASGGGGGDCCV 321
G D+ +++W++ + + + + H V +++ P + N++ASG G D V
Sbjct: 351 VGGSEDHGIYLWELQT------RKIVQKLEGHSDTVVSVSCHPTE-NMIASGALGNDKTV 509
Query: 322 KLW 324
K+W
Sbjct: 510 KIW 518
Score = 44.7 bits (104), Expect = 3e-05
Identities = 36/169 (21%), Positives = 65/169 (38%), Gaps = 2/169 (1%)
Frame = +3
Query: 245 HRREVCGLKWSLDGKQLASGGNDNVVHIWDMSAVSSNSPTRWLYRFDEHKAAVKALAWCP 304
H V + ++ DG + S D + IWD S D+ V + + P
Sbjct: 36 HSDPVTAVDFNRDGSLIVSSSYDGLCRIWDASTGHCMKTL-----IDDENPPVSFVKFSP 200
Query: 305 FQGNLLASGGGGGDCCVKLWNTGMGERMNSVD--TGSQVCALLWSKNERELLSSHGLTQN 362
+L G D ++LWN G + + S+ C G +
Sbjct: 201 NAKFILV---GTLDNNLRLWNYSTGRFLKTYTGHVNSKYCISSTFSTTNGKYVVGGSEDH 371
Query: 363 QLTLWKYPSMLKIAELHGHTSRVLHMTQSPDGSTVASAAAAADQTLRFW 411
+ LW+ + + +L GH+ V+ ++ P + +AS A D+T++ W
Sbjct: 372 GIYLWELQTRKIVQKLEGHSDTVVSVSCHPTENMIASGALGNDKTVKIW 518
>TC11756 weakly similar to GB|AAH01494.1|16306637|BC001494 U5 snRNP-specific
40 kDa protein (hPrp8-binding) {Homo sapiens;} , partial
(43%)
Length = 719
Score = 48.1 bits (113), Expect = 3e-06
Identities = 32/125 (25%), Positives = 56/125 (44%), Gaps = 2/125 (1%)
Frame = +1
Query: 203 GHRARVGSLAWN--GHVLTTGGMDGKIVNNDVRLRSQIINTYRGHRREVCGLKWSLDGKQ 260
GH++ + ++ +N G V+ +G D +I +V + +GH+ V L W+ DG Q
Sbjct: 298 GHQSVIYTMKFNPAGTVIASGSHDREIFLWNVHGECKNFMVLKGHKNAVLDLHWTTDGTQ 477
Query: 261 LASGGNDNVVHIWDMSAVSSNSPTRWLYRFDEHKAAVKALAWCPFQGNLLASGGGGGDCC 320
+ S D V +WD+ + + + EH + V + CP + G D
Sbjct: 478 IVSASPDKTVRVWDVET------GKQVKKMVEHLSYVNSC--CPSRRGPPLVVSGSDDGT 633
Query: 321 VKLWN 325
KLW+
Sbjct: 634 AKLWD 648
Score = 32.7 bits (73), Expect = 0.12
Identities = 22/78 (28%), Positives = 35/78 (44%), Gaps = 1/78 (1%)
Frame = +1
Query: 337 TGSQVCALLWSKNERELLSSHGLTQNQLTLWKYPSMLK-IAELHGHTSRVLHMTQSPDGS 395
TG Q N + + G ++ LW K L GH + VL + + DG+
Sbjct: 295 TGHQSVIYTMKFNPAGTVIASGSHDREIFLWNVHGECKNFMVLKGHKNAVLDLHWTTDGT 474
Query: 396 TVASAAAAADQTLRFWEV 413
+ S A+ D+T+R W+V
Sbjct: 475 QIVS--ASPDKTVRVWDV 522
Score = 28.1 bits (61), Expect = 3.0
Identities = 33/136 (24%), Positives = 55/136 (40%), Gaps = 8/136 (5%)
Frame = +1
Query: 148 SDSSGSEFVTVDEEEGPVTSVCWAPDGRHLAVGLTNSHVQLWDTAAN-KQLRTLKGGHRA 206
+ S S + + + + ++ + P G +A G + + LW+ K LKG H+
Sbjct: 262 TSSLESPIMLLTGHQSVIYTMKFNPAGTVIASGSHDREIFLWNVHGECKNFMVLKG-HKN 438
Query: 207 RVGSLAWNGHVLTTGGMDGKIVNNDVRLRSQIINTYRGHRREVCGLKW------SLDGKQ 260
V L W TT G + D +R + T + ++ V L + S G
Sbjct: 439 AVLDLHW-----TTDGTQIVSASPDKTVRVWDVETGKQVKKMVEHLSYVNSCCPSRRGPP 603
Query: 261 L-ASGGNDNVVHIWDM 275
L SG +D +WDM
Sbjct: 604 LVVSGSDDGTAKLWDM 651
>AV407645
Length = 413
Score = 47.4 bits (111), Expect = 5e-06
Identities = 36/145 (24%), Positives = 66/145 (44%), Gaps = 11/145 (7%)
Frame = +2
Query: 245 HRREVCGLKWSLDGKQLASGGNDNVVHIWDMSAVSSNSPTRWLYRFDEHKAAVKALAWCP 304
H EV +++S +GK LAS ND IW++ + +R H+ A +++W P
Sbjct: 5 HDDEVWLVQFSHNGKYLASASNDRTAIIWEVGI----NGLSLKHRLSGHQKAFSSISWSP 172
Query: 305 FQGNLLASGGGGGDCCVKLWNTGMGERMNSVD-TGSQVCALLWSKNERELLSSHGLTQNQ 363
LL G + ++ W+ G+ + + G+ + + W + + +LS GL+
Sbjct: 173 NDQELLTC---GVEEAIRRWDVSTGKCLQIYEKAGAGLISCTWFPSGKYILS--GLSDKS 337
Query: 364 LTL----------WKYPSMLKIAEL 378
+ + WK P LKI++L
Sbjct: 338 ICMWELDGKEVESWKGPKTLKISDL 412
Score = 30.8 bits (68), Expect = 0.46
Identities = 22/109 (20%), Positives = 46/109 (42%), Gaps = 1/109 (0%)
Frame = +2
Query: 214 NGHVLTTGGMDGKIVNNDVRLRS-QIINTYRGHRREVCGLKWSLDGKQLASGGNDNVVHI 272
NG L + D + +V + + + GH++ + WS + ++L + G + +
Sbjct: 41 NGKYLASASNDRTAIIWEVGINGLSLKHRLSGHQKAFSSISWSPNDQELLTCGVEEAIRR 220
Query: 273 WDMSAVSSNSPTRWLYRFDEHKAAVKALAWCPFQGNLLASGGGGGDCCV 321
WD+S + L +++ A + + W P G + SG C+
Sbjct: 221 WDVST------GKCLQIYEKAGAGLISCTWFP-SGKYILSGLSDKSICM 346
Score = 28.9 bits (63), Expect = 1.7
Identities = 30/157 (19%), Positives = 57/157 (36%)
Frame = +2
Query: 119 DDFSLNLLDWGSRNVLSIALDHTIYFWNASDSSGSEFVTVDEEEGPVTSVCWAPDGRHLA 178
D+ L + + S + D T W + S + + +S+ W+P+ + L
Sbjct: 11 DEVWLVQFSHNGKYLASASNDRTAIIWEVGINGLSLKHRLSGHQKAFSSISWSPNDQELL 190
Query: 179 VGLTNSHVQLWDTAANKQLRTLKGGHRARVGSLAWNGHVLTTGGMDGKIVNNDVRLRSQI 238
++ WD + K L ++ A G + T GK + + + +S
Sbjct: 191 TCGVEEAIRRWDVSTGKCL---------QIYEKAGAGLISCTWFPSGKYILSGLSDKS-- 337
Query: 239 INTYRGHRREVCGLKWSLDGKQLASGGNDNVVHIWDM 275
+C W LDGK++ S + I D+
Sbjct: 338 ----------IC--MWELDGKEVESWKGPKTLKISDL 412
>TC16077 similar to PIR|T00593|T02480 sec13-related protein At2g30050 -
Arabidopsis thaliana {Arabidopsis thaliana;}, partial
(46%)
Length = 521
Score = 46.2 bits (108), Expect = 1e-05
Identities = 46/151 (30%), Positives = 64/151 (41%), Gaps = 13/151 (8%)
Frame = +1
Query: 184 SHVQLWDTAANKQLRTLKG-----GHRARVGSLAWN--GHVLTTGGMDGKIVNNDVRLR- 235
+H QL T R + G GH+ V +A + G L T D I V +
Sbjct: 46 THCQLHLTRLTPHRRNMPGQKVETGHQDTVHDVAMDYYGKRLATASSDHTIKIIGVSIAA 225
Query: 236 SQIINTYRGHRREVCGLKWSLD--GKQLASGGNDNVVHIWDMSAVSSNSPTRWL--YRFD 291
SQ + T GH+ V + W+ G LAS D V +W W + FD
Sbjct: 226 SQHLATLTGHQGPVWQVAWAHPKFGSLLASCSYDGRVILW-----KEGDQNEWTQAHVFD 390
Query: 292 EHKAAVKALAWCPFQ-GNLLASGGGGGDCCV 321
EHK++V ++AW P + G LA G G+ V
Sbjct: 391 EHKSSVNSVAWAPHELGLCLACGSSDGNISV 483
Score = 40.0 bits (92), Expect = 8e-04
Identities = 30/110 (27%), Positives = 46/110 (41%), Gaps = 8/110 (7%)
Frame = +1
Query: 126 LDWGSRNVLSIALDHTIYFWNASDSSGSEFVTVDEEEGPVTSVCWAPD--GRHLAVGLTN 183
+D+ + + + + DHTI S ++ T+ +GPV V WA G LA +
Sbjct: 148 MDYYGKRLATASSDHTIKIIGVSIAASQHLATLTGHQGPVWQVAWAHPKFGSLLASCSYD 327
Query: 184 SHVQLWDTAANKQLRT--LKGGHRARVGSLAWNGH----VLTTGGMDGKI 227
V LW + + H++ V S+AW H L G DG I
Sbjct: 328 GRVILWKEGDQNEWTQAHVFDEHKSSVNSVAWAPHELGLCLACGSSDGNI 477
>TC13796 homologue to UP|Q9M7I2 (Q9M7I2) WD-repeat cell cycle regulatory
protein, partial (13%)
Length = 529
Score = 45.1 bits (105), Expect = 2e-05
Identities = 21/39 (53%), Positives = 28/39 (70%)
Frame = +3
Query: 376 AELHGHTSRVLHMTQSPDGSTVASAAAAADQTLRFWEVF 414
A L GH+ RVL+++ SPDG T+ + A D+TLRFW VF
Sbjct: 3 ATLTGHSYRVLYLSISPDGQTIVT--GAGDETLRFWNVF 113
Score = 26.6 bits (57), Expect = 8.6
Identities = 11/35 (31%), Positives = 17/35 (48%)
Frame = +3
Query: 241 TYRGHRREVCGLKWSLDGKQLASGGNDNVVHIWDM 275
T GH V L S DG+ + +G D + W++
Sbjct: 6 TLTGHSYRVLYLSISPDGQTIVTGAGDETLRFWNV 110
>TC8856 similar to UP|Q8W403 (Q8W403) Sec13p, partial (48%)
Length = 568
Score = 44.7 bits (104), Expect = 3e-05
Identities = 41/143 (28%), Positives = 63/143 (43%), Gaps = 8/143 (5%)
Frame = +1
Query: 187 QLWDTAANKQLRTLKGGHRARVGSLAWN--GHVLTTGGMDGKIVNNDV-RLRSQIINTYR 243
QL+ N + ++ GH+ V +A + G L T D I V SQ + T
Sbjct: 94 QLFIIHRNMPAQKVETGHQDTVHDVAMDYYGKRLATASSDHTIKIIGVSNTASQHLATLA 273
Query: 244 GHRREVCGLKWSLD--GKQLASGGNDNVVHIWDMSAVSSNSPTRWL--YRFDEHKAAVKA 299
GH+ V + W+ G +AS D V IW + W+ + FD+HK++V +
Sbjct: 274 GHQGPVWQVAWAHPKFGSMIASCSYDGRVIIW-----KEGNQNEWIQAHVFDDHKSSVNS 438
Query: 300 LAWCPFQ-GNLLASGGGGGDCCV 321
+ W P + G LA G G+ V
Sbjct: 439 VVWAPHELGLCLACGSSDGNISV 507
Score = 38.9 bits (89), Expect = 0.002
Identities = 28/110 (25%), Positives = 50/110 (45%), Gaps = 8/110 (7%)
Frame = +1
Query: 126 LDWGSRNVLSIALDHTIYFWNASDSSGSEFVTVDEEEGPVTSVCWAPD--GRHLAVGLTN 183
+D+ + + + + DHTI S+++ T+ +GPV V WA G +A +
Sbjct: 172 MDYYGKRLATASSDHTIKIIGVSNTASQHLATLAGHQGPVWQVAWAHPKFGSMIASCSYD 351
Query: 184 SHVQLW-DTAANKQLRT-LKGGHRARVGSLAWNGH----VLTTGGMDGKI 227
V +W + N+ ++ + H++ V S+ W H L G DG I
Sbjct: 352 GRVIIWKEGNQNEWIQAHVFDDHKSSVNSVVWAPHELGLCLACGSSDGNI 501
>AV413963
Length = 313
Score = 43.9 bits (102), Expect = 5e-05
Identities = 25/84 (29%), Positives = 38/84 (44%)
Frame = +2
Query: 242 YRGHRREVCGLKWSLDGKQLASGGNDNVVHIWDMSAVSSNSPTRWLYRFDEHKAAVKALA 301
Y GH+++V + W+ G +LASG D IW + H +V L
Sbjct: 65 YSGHKKKVHSVAWNCIGTKLASGSVDQTARIWHVEQHGHGKVKD--IELKGHTDSVDQLC 238
Query: 302 WCPFQGNLLASGGGGGDCCVKLWN 325
W P +L+A+ GD V+LW+
Sbjct: 239 WDPKHADLIAT--ASGDKTVRLWD 304
Score = 30.4 bits (67), Expect = 0.60
Identities = 23/78 (29%), Positives = 37/78 (46%), Gaps = 6/78 (7%)
Frame = +2
Query: 203 GHRARVGSLAWN--GHVLTTGGMD--GKIVNNDVRLRSQIIN-TYRGHRREVCGLKWSLD 257
GH+ +V S+AWN G L +G +D +I + + ++ + +GH V L W
Sbjct: 71 GHKKKVHSVAWNCIGTKLASGSVDQTARIWHVEQHGHGKVKDIELKGHTDSVDQLCWDPK 250
Query: 258 GKQL-ASGGNDNVVHIWD 274
L A+ D V +WD
Sbjct: 251 HADLIATASGDKTVRLWD 304
>TC8529 homologue to UP|Q94KS2 (Q94KS2) TGF-beta receptor-interacting
protein 1, partial (44%)
Length = 650
Score = 43.1 bits (100), Expect = 9e-05
Identities = 32/130 (24%), Positives = 53/130 (40%), Gaps = 16/130 (12%)
Frame = +1
Query: 159 DEEEG---PVTSVCWAPDGRHLAVGLTNSHVQLWDTAANKQLRTLKGGHRARVGSLA-WN 214
D+E G +TS+ + DG H G + +LWDT ++T +++
Sbjct: 16 DKESGHKKAITSLSKSADGSHFVTGSLDKSARLWDTRTLTLIKTYVTERPVNAVAMSPLL 195
Query: 215 GHVLTTGGMDGKIVNND------------VRLRSQIINTYRGHRREVCGLKWSLDGKQLA 262
HV+ GG D V ++ + I +GH + L ++ DGK +
Sbjct: 196 DHVVLGGGQDASAVTTTDHRAGKFEAKFYDKILQEEIGGVKGHFGPINALAFNPDGKSFS 375
Query: 263 SGGNDNVVHI 272
SGG D V +
Sbjct: 376 SGGEDGYVRL 405
Score = 30.8 bits (68), Expect = 0.46
Identities = 18/71 (25%), Positives = 30/71 (41%)
Frame = +1
Query: 244 GHRREVCGLKWSLDGKQLASGGNDNVVHIWDMSAVSSNSPTRWLYRFDEHKAAVKALAWC 303
GH++ + L S DG +G D +WD + T L + + V A+A
Sbjct: 28 GHKKAITSLSKSADGSHFVTGSLDKSARLWD-------TRTLTLIKTYVTERPVNAVAMS 186
Query: 304 PFQGNLLASGG 314
P +++ GG
Sbjct: 187 PLLDHVVLGGG 219
>TC15844 homologue to UP|Q9FV61 (Q9FV61) Heterotrimeric GTP-binding protein
subunit beta 1, partial (28%)
Length = 668
Score = 40.4 bits (93), Expect = 6e-04
Identities = 28/89 (31%), Positives = 40/89 (44%), Gaps = 5/89 (5%)
Frame = +3
Query: 164 PVTSVCWAPDGRHLAVGLTNSHVQLWDTAANK---QLRTLKGGHRARVG--SLAWNGHVL 218
PVTS+ ++ GR L G N +WDT K + +L+ H R+ L+ +G L
Sbjct: 90 PVTSIAFSISGRLLFAGYANGDCYVWDTLLAKVVLNIGSLQDSHEGRISCLGLSADGSAL 269
Query: 219 TTGGMDGKIVNNDVRLRSQIINTYRGHRR 247
TGG D + I + GHRR
Sbjct: 270 CTGGWDTNLK----------IWAFGGHRR 326
Score = 26.6 bits (57), Expect = 8.6
Identities = 12/52 (23%), Positives = 26/52 (49%)
Frame = +3
Query: 159 DEEEGPVTSVCWAPDGRHLAVGLTNSHVQLWDTAANKQLRTLKGGHRARVGS 210
D EG ++ + + DG L G ++++++W +++ +GG GS
Sbjct: 213 DSHEGRISCLGLSADGSALCTGGWDTNLKIWAFGGHRRT*AKEGGKHLSYGS 368
>TC10816 homologue to UP|CAE45585 (CAE45585) Coatomer alpha subunit-like
protein, partial (20%)
Length = 979
Score = 39.3 bits (90), Expect = 0.001
Identities = 36/148 (24%), Positives = 63/148 (42%), Gaps = 7/148 (4%)
Frame = +1
Query: 273 WDMSAVSSNSPTRWLYR-------FDEHKAAVKALAWCPFQGNLLASGGGGGDCCVKLWN 325
W ++++ S W YR FDEH V+ + + Q ++ GG D +K+WN
Sbjct: 307 WILASLHSGVIQLWDYRMGTLIDKFDEHDGPVRGVHFHHSQPLFVS---GGDDYKIKVWN 477
Query: 326 TGMGERMNSVDTGSQVCALLWSKNERELLSSHGLTQNQLTLWKYPSMLKIAELHGHTSRV 385
+ + ++ + +E + S Q + +W + S I+ L GH V
Sbjct: 478 YKLHRCLFTLLGHLDYIRTVQFHHESPWIVSASDDQT-IRIWNWQSRTCISVLTGHNHYV 654
Query: 386 LHMTQSPDGSTVASAAAAADQTLRFWEV 413
+ P V SA+ DQT+R W++
Sbjct: 655 MCALFHPREDLVVSASL--DQTVRVWDI 732
>AV780136
Length = 558
Score = 38.5 bits (88), Expect = 0.002
Identities = 31/106 (29%), Positives = 50/106 (46%), Gaps = 2/106 (1%)
Frame = -1
Query: 210 SLAWN--GHVLTTGGMDGKIVNNDVRLRSQIINTYRGHRREVCGLKWSLDGKQLASGGND 267
+LA N G VL +GG + I D R S+ + RGH + L G+ SG +D
Sbjct: 558 ALAMNEGGTVLVSGGTEKVIRVWDPRSGSKTLKL-RGHADNIRALLLDSTGRYCLSGSSD 382
Query: 268 NVVHIWDMSAVSSNSPTRWLYRFDEHKAAVKALAWCPFQGNLLASG 313
+++ +WD+ R L+ + H +V ALA P ++ + G
Sbjct: 381 SMIRLWDI------GQPRCLHTYAVHTDSVWALASTPTFSHVYSGG 262
Score = 30.0 bits (66), Expect = 0.78
Identities = 26/110 (23%), Positives = 48/110 (43%), Gaps = 2/110 (1%)
Frame = -1
Query: 174 GRHLAVGLTNSHVQLWDTAANKQLRTLKGGHRARVGSLAWN--GHVLTTGGMDGKIVNND 231
G L G T +++WD + + L+G H + +L + G +G D I D
Sbjct: 537 GTVLVSGGTEKVIRVWDPRSGSKTLKLRG-HADNIRALLLDSTGRYCLSGSSDSMIRLWD 361
Query: 232 VRLRSQIINTYRGHRREVCGLKWSLDGKQLASGGNDNVVHIWDMSAVSSN 281
+ + + ++TY H V L + + SGG D +++ D+ S+
Sbjct: 360 IG-QPRCLHTYAVHTDSVWALASTPTFSHVYSGGRDFSLYLTDLQTRESS 214
Score = 28.1 bits (61), Expect = 3.0
Identities = 35/149 (23%), Positives = 63/149 (41%), Gaps = 4/149 (2%)
Frame = -1
Query: 129 GSRNVLSIALDHTIYFWNASDSSGSEFVTVDEEEGPVTSVCWAPDGRHLAVGLTNSHVQL 188
G ++S + I W+ SGS+ + + + ++ GR+ G ++S ++L
Sbjct: 540 GGTVLVSGGTEKVIRVWDPR--SGSKTLKLRGHADNIRALLLDSTGRYCLSGSSDSMIRL 367
Query: 189 WDTAANKQLRTLKGGHRARVGSLAWN---GHVLTTGGMDGKIVNNDVRLR-SQIINTYRG 244
WD + L T H V +LA HV +GG D + D++ R S ++ T
Sbjct: 366 WDIGQPRCLHTY-AVHTDSVWALASTPTFSHVY-SGGRDFSLYLTDLQTRESSLLCTGEH 193
Query: 245 HRREVCGLKWSLDGKQLASGGNDNVVHIW 273
R++ +L + D+ VH W
Sbjct: 192 PIRQL-----ALHDDSIWVASTDSSVHKW 121
>TC14937 similar to UP|Q9FN19 (Q9FN19) Genomic DNA, chromosome 5, TAC
clone:K8K14 (AT5g67320/K8K14_4), partial (19%)
Length = 1089
Score = 37.7 bits (86), Expect = 0.004
Identities = 20/59 (33%), Positives = 32/59 (53%)
Frame = +1
Query: 217 VLTTGGMDGKIVNNDVRLRSQIINTYRGHRREVCGLKWSLDGKQLASGGNDNVVHIWDM 275
VL + D + DV + ++I + GHR V + +S +G+ LASG D +HIW +
Sbjct: 52 VLASASFDSTVKLWDVEV-GKLIYSLNGHRDGVYSVAFSPNGEYLASGSPDKSIHIWSL 225
Score = 35.0 bits (79), Expect = 0.024
Identities = 19/80 (23%), Positives = 36/80 (44%)
Frame = +1
Query: 135 SIALDHTIYFWNASDSSGSEFVTVDEEEGPVTSVCWAPDGRHLAVGLTNSHVQLWDTAAN 194
S + D T+ W+ G +++ V SV ++P+G +LA G + + +W
Sbjct: 61 SASFDSTVKLWDVE--VGKLIYSLNGHRDGVYSVAFSPNGEYLASGSPDKSIHIWSLKEG 234
Query: 195 KQLRTLKGGHRARVGSLAWN 214
K ++T G + + WN
Sbjct: 235 KIVKTYTGS--GGIFEVCWN 288
Score = 27.3 bits (59), Expect = 5.1
Identities = 16/67 (23%), Positives = 31/67 (45%)
Frame = +1
Query: 347 SKNERELLSSHGLTQNQLTLWKYPSMLKIAELHGHTSRVLHMTQSPDGSTVASAAAAADQ 406
S ++L+ + + + LW I L+GH V + SP+G +AS + D+
Sbjct: 31 SNPNKKLVLASASFDSTVKLWDVEVGKLIYSLNGHRDGVYSVAFSPNGEYLAS--GSPDK 204
Query: 407 TLRFWEV 413
++ W +
Sbjct: 205 SIHIWSL 225
>TC19491 similar to GB|AAP37692.1|30725340|BT008333 At4g29830 {Arabidopsis
thaliana;}, partial (39%)
Length = 527
Score = 37.0 bits (84), Expect = 0.006
Identities = 24/81 (29%), Positives = 41/81 (49%), Gaps = 3/81 (3%)
Frame = +1
Query: 203 GHRARVGSLAWNGH---VLTTGGMDGKIVNNDVRLRSQIINTYRGHRREVCGLKWSLDGK 259
GH V SL ++ + +L T DG + D ++ ++ T GH V + S DG
Sbjct: 10 GHFMPVRSLVYSPYDPRLLFTASDDGNVHMYDAEGKA-LVGTMSGHASWVLCVDVSPDGG 186
Query: 260 QLASGGNDNVVHIWDMSAVSS 280
+A+G +D V +WD++ +S
Sbjct: 187 AIATGSSDRTVRLWDLAMRAS 249
Score = 32.7 bits (73), Expect = 0.12
Identities = 14/39 (35%), Positives = 25/39 (63%)
Frame = +1
Query: 375 IAELHGHTSRVLHMTQSPDGSTVASAAAAADQTLRFWEV 413
+ + GH S VL + SPDG +A+ ++D+T+R W++
Sbjct: 124 VGTMSGHASWVLCVDVSPDGGAIAT--GSSDRTVRLWDL 234
Score = 30.0 bits (66), Expect = 0.78
Identities = 20/66 (30%), Positives = 33/66 (49%), Gaps = 1/66 (1%)
Frame = +1
Query: 149 DSSGSEFV-TVDEEEGPVTSVCWAPDGRHLAVGLTNSHVQLWDTAANKQLRTLKGGHRAR 207
D+ G V T+ V V +PDG +A G ++ V+LWD A ++T+ H +
Sbjct: 103 DAEGKALVGTMSGHASWVLCVDVSPDGGAIATGSSDRTVRLWDLAMRASVQTM-SNHTDQ 279
Query: 208 VGSLAW 213
V +A+
Sbjct: 280 VWGVAF 297
>TC19190 weakly similar to PIR|T46032|T46032 WD-40 repeat regulatory protein
tup1 homolog - Arabidopsis thaliana
{Arabidopsis thaliana;}, partial (18%)
Length = 511
Score = 37.0 bits (84), Expect = 0.006
Identities = 17/46 (36%), Positives = 24/46 (51%)
Frame = +3
Query: 366 LWKYPSMLKIAELHGHTSRVLHMTQSPDGSTVASAAAAADQTLRFW 411
LW I +L GHT V+ +T P + +ASA D+T+R W
Sbjct: 57 LWDLQQKNMIQKLEGHTDTVISVTCHPTENKIASAGLDGDRTVRVW 194
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.318 0.133 0.424
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,421,662
Number of Sequences: 28460
Number of extensions: 153554
Number of successful extensions: 1807
Number of sequences better than 10.0: 108
Number of HSP's better than 10.0 without gapping: 1560
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1751
length of query: 436
length of database: 4,897,600
effective HSP length: 93
effective length of query: 343
effective length of database: 2,250,820
effective search space: 772031260
effective search space used: 772031260
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 57 (26.6 bits)
Medicago: description of AC146343.9


