

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146343.4 - phase: 0
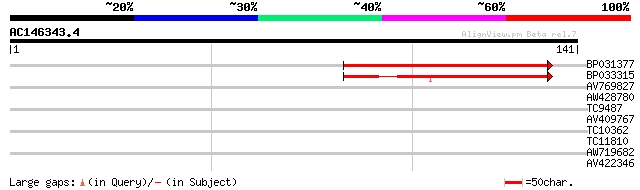
(141 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BP031377 85 5e-18
BP033315 56 2e-09
AV769827 35 0.005
AW428780 33 0.014
TC9487 homologue to UP|BAC99050 (BAC99050) Brassinosteroid recep... 25 3.9
AV409767 25 3.9
TC10362 similar to UP|Q8L5A7 (Q8L5A7) Steroid sulfotransferase-l... 25 3.9
TC11810 25 6.6
AW719682 25 6.6
AV422346 24 8.6
>BP031377
Length = 401
Score = 84.7 bits (208), Expect = 5e-18
Identities = 41/52 (78%), Positives = 44/52 (83%)
Frame = -3
Query: 84 DFIIVGRRNGIKSPQTQALASWTEYPELGVLGDLLASPDTNTKASILVVQQQ 135
DFIIVGRRNG+KS T L +WTEY ELGV+GDLLASPD TKASILVVQQQ
Sbjct: 390 DFIIVGRRNGVKSEMTAGLENWTEYTELGVIGDLLASPDMETKASILVVQQQ 235
>BP033315
Length = 433
Score = 56.2 bits (134), Expect = 2e-09
Identities = 31/57 (54%), Positives = 39/57 (68%), Gaps = 5/57 (8%)
Frame = -1
Query: 84 DFIIVGRRNGIKSPQTQALA-----SWTEYPELGVLGDLLASPDTNTKASILVVQQQ 135
D ++VGR + PQT ++ W+E PELG +GD+LASPD TKASILVVQQQ
Sbjct: 352 DLVMVGREH----PQTPSVLLKGHEKWSECPELGTIGDMLASPDFVTKASILVVQQQ 194
>AV769827
Length = 438
Score = 35.0 bits (79), Expect = 0.005
Identities = 20/53 (37%), Positives = 31/53 (57%), Gaps = 1/53 (1%)
Frame = -2
Query: 83 HDFIIVGRRNGIKSPQTQALASWTEYPELGVLGDLLASPDTN-TKASILVVQQ 134
+D IIVG+ + + EY ELG +GD+LAS + ++S+LV+QQ
Sbjct: 374 YDLIIVGKGRFPSTMVAELAERHAEYAELGPVGDILASSTGHKMRSSVLVIQQ 216
>AW428780
Length = 330
Score = 33.5 bits (75), Expect = 0.014
Identities = 20/54 (37%), Positives = 30/54 (55%), Gaps = 2/54 (3%)
Frame = +2
Query: 82 QHDFIIVGRRNGI--KSPQTQALASWTEYPELGVLGDLLASPDTNTKASILVVQ 133
+++FIIVG+ I + S E+ ELG +GDLL S +S+LV+Q
Sbjct: 86 EYEFIIVGKGQQILESTMMLNIQDSRLEHAELGPVGDLLTSSGQGITSSVLVIQ 247
>TC9487 homologue to UP|BAC99050 (BAC99050) Brassinosteroid receptor,
partial (8%)
Length = 744
Score = 25.4 bits (54), Expect = 3.9
Identities = 11/16 (68%), Positives = 13/16 (80%)
Frame = -2
Query: 86 IIVGRRNGIKSPQTQA 101
IIVGRR+G+ S Q QA
Sbjct: 140 IIVGRRHGLSSKQAQA 93
>AV409767
Length = 411
Score = 25.4 bits (54), Expect = 3.9
Identities = 13/29 (44%), Positives = 18/29 (61%), Gaps = 1/29 (3%)
Frame = -2
Query: 15 KILITWVVYHLVSTIKN-DEFTSWDVMLD 42
KI +VYHL+ I+N EFTS + +D
Sbjct: 335 KIRQNTIVYHLIMQIRNLTEFTSKTISID 249
>TC10362 similar to UP|Q8L5A7 (Q8L5A7) Steroid sulfotransferase-like protein
(At5g07010), partial (23%)
Length = 425
Score = 25.4 bits (54), Expect = 3.9
Identities = 11/24 (45%), Positives = 14/24 (57%)
Frame = -3
Query: 5 HCVWLKGQSKKILITWVVYHLVST 28
HC WL S+K L W+ + LV T
Sbjct: 237 HCAWLGSASEKCL--WLGFQLVGT 172
>TC11810
Length = 640
Score = 24.6 bits (52), Expect = 6.6
Identities = 23/85 (27%), Positives = 34/85 (39%), Gaps = 16/85 (18%)
Frame = +1
Query: 16 ILITWVVYHLVSTIKNDEFTSWDVMLDDEL--------------LKGVKGVYGSVDNVTY 61
+L W+V V + D+ + V LDDE LKG ++D V
Sbjct: 136 LLSDWIVDANVQSFDGDKNYPFGVELDDEYETDSIDYDDGAARPLKGSSLELETMDGVAV 315
Query: 62 EKVEVE--NTSDTTEFISDIAIQHD 84
+VE N D ++ SD+ HD
Sbjct: 316 GSPDVEHANIDDASDDESDLNYFHD 390
>AW719682
Length = 480
Score = 24.6 bits (52), Expect = 6.6
Identities = 14/35 (40%), Positives = 21/35 (60%)
Frame = +1
Query: 30 KNDEFTSWDVMLDDELLKGVKGVYGSVDNVTYEKV 64
++ EF S + +DD+L KG+KGV T +KV
Sbjct: 208 RSPEFIS--LFVDDKLRKGLKGVSEDDIETTLDKV 306
>AV422346
Length = 465
Score = 24.3 bits (51), Expect = 8.6
Identities = 6/18 (33%), Positives = 11/18 (60%)
Frame = +3
Query: 6 CVWLKGQSKKILITWVVY 23
C+WL+ K ++ W +Y
Sbjct: 153 CLWLESMRKHTILAWTLY 206
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.316 0.133 0.387
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,291,683
Number of Sequences: 28460
Number of extensions: 23592
Number of successful extensions: 145
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 145
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 145
length of query: 141
length of database: 4,897,600
effective HSP length: 81
effective length of query: 60
effective length of database: 2,592,340
effective search space: 155540400
effective search space used: 155540400
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 51 (24.3 bits)
Medicago: description of AC146343.4


