

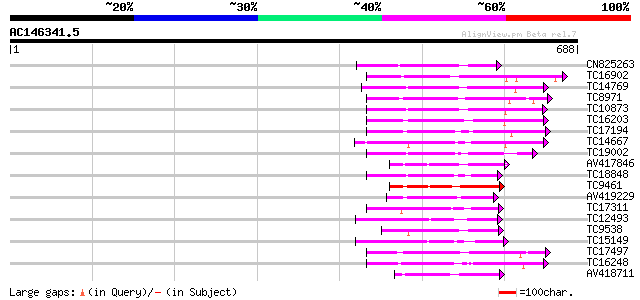
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146341.5 + phase: 0 /pseudo
(688 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CN825263 133 8e-32
TC16902 homologue to UP|Q8VZH5 (Q8VZH5) Receptor protein kinase-... 128 3e-30
TC14769 UP|Q8LKX1 (Q8LKX1) Receptor-like kinase SYMRK, complete 125 2e-29
TC8971 similar to GB|AAM16251.1|20334780|AY093990 At2g11520/F14P... 124 5e-29
TC10873 similar to UP|Q9LDZ5 (Q9LDZ5) F2D10.13 (F5M15.3), partia... 124 5e-29
TC16203 UP|Q8GRU6 (Q8GRU6) LRR receptor-like kinase (Hypernodula... 122 2e-28
TC17194 UP|CAE02590 (CAE02590) Nod-factor receptor 1b, complete 117 8e-27
TC14667 homologue to UP|Q84P43 (Q84P43) Protein kinase Pti1, com... 115 2e-26
TC19002 weakly similar to UP|Q9XJ10 (Q9XJ10) ESTs C22458(C62866)... 105 2e-23
AV417846 105 2e-23
TC18848 weakly similar to UP|Q9SSQ6 (Q9SSQ6) F6D8.24 protein, pa... 105 3e-23
TC9461 similar to UP|O49840 (O49840) Protein kinase (AT2G02800/T... 104 4e-23
AV419229 103 9e-23
TC17311 similar to UP|Q8K2Y2 (Q8K2Y2) Receptor-interacting serin... 103 9e-23
TC12493 similar to UP|Q9LP77 (Q9LP77) T1N15.9, partial (35%) 100 8e-22
TC9538 similar to UP|RLK5_ARATH (P47735) Receptor-like protein k... 97 7e-21
TC15149 similar to UP|Q9LVI6 (Q9LVI6) Probable receptor-like pro... 97 1e-20
TC17497 UP|CAE45595 (CAE45595) S-receptor kinase-like protein 2,... 95 3e-20
TC16248 similar to UP|BAC87845 (BAC87845) Leucine-rich repeat re... 95 4e-20
AV418711 94 6e-20
>CN825263
Length = 663
Score = 133 bits (335), Expect = 8e-32
Identities = 73/178 (41%), Positives = 102/178 (57%), Gaps = 2/178 (1%)
Frame = +1
Query: 421 KDPQNFNRSLSYQVEVLSKLKHPNLITLIGVNQESKT--LIYEYLPNGSLEDHLSRNGNN 478
+D Q R +VE+LS+L H NL+ LIG+ E +T LIYE +PNGS+E HL +
Sbjct: 67 RDDQRGGREFLAEVEMLSRLHHRNLVKLIGICIEKQTRCLIYELVPNGSVESHL-HGADK 243
Query: 479 NAPPLTWQTRIRIATELCSALIFLHSNKPHSIVHGDLKPSNILLDANLVTKLSDFGICRV 538
PL W R++IA L +LH + ++H D K SNILL+ + K+SDFG+ R
Sbjct: 244 ETGPLDWNARMKIALGAARGLAYLHEDSNPCVIHRDFKSSNILLECDFTPKVSDFGLART 423
Query: 539 LSCQNDSSSNNSTTQFWITSFAKGTFAYMDPEFLGTGELTSKSDVYSFGIVLLRLITG 596
+ + I++ GTF Y+ PE+ TG L KSDVYS+G+VLL L+TG
Sbjct: 424 ALDEGNKH---------ISTHVMGTFGYLAPEYAMTGHLLVKSDVYSYGVVLLELLTG 570
>TC16902 homologue to UP|Q8VZH5 (Q8VZH5) Receptor protein kinase-like
protein, partial (46%)
Length = 941
Score = 128 bits (322), Expect = 3e-30
Identities = 83/257 (32%), Positives = 135/257 (52%), Gaps = 13/257 (5%)
Frame = +3
Query: 433 QVEVLSKLKHPNLITLIGVNQESKT--LIYEYLPNGSLEDHLSRNGNNNAPPLTWQTRIR 490
++E+LSKL+H +L++LIG +E+ L+Y+++ G+L +HL + PPL W+ R+
Sbjct: 129 EIEMLSKLRHRHLVSLIGYCEENTEMILVYDHMAYGTLREHLYKT---QKPPLPWKQRLE 299
Query: 491 IATELCSALIFLHSNKPHSIVHGDLKPSNILLDANLVTKLSDFGICRVLSCQNDSSSNNS 550
I L +LH+ ++I+H D+K +NILLD V K+SDFG+ S + +
Sbjct: 300 ICIGAARGLHYLHTGAKYTIIHRDVKTTNILLDEKWVAKVSDFGL---------SKTGPT 452
Query: 551 TTQFWITSFAKGTFAYMDPEFLGTGELTSKSDVYSFGIVLLRLITGKPALG---IKNEVL 607
+++ KG+F Y+DPE+ +LT KSDVYSFG+VL ++ +PAL K +V
Sbjct: 453 LDNTHVSTVVKGSFGYLDPEYFRRQQLTDKSDVYSFGVVLFEILCARPALNPSLAKEQVS 632
Query: 608 YALNNA----GGNVKSVLDP-LAGDWPIVEAEKLVHFALRCCDMNKKSRPELCSEGWRV- 661
A + G + +LDP L G +K A++C RP + W +
Sbjct: 633 LAEWASHCYNKGILDQILDPYLKGKIAPECFKKFAETAMKCVSDQGIERPSMGDVLWNLE 812
Query: 662 --LEPMKVSCSGTNNFG 676
L+ + + N FG
Sbjct: 813 FALQLQESAEESGNGFG 863
>TC14769 UP|Q8LKX1 (Q8LKX1) Receptor-like kinase SYMRK, complete
Length = 3495
Score = 125 bits (314), Expect = 2e-29
Identities = 77/237 (32%), Positives = 126/237 (52%), Gaps = 10/237 (4%)
Frame = +3
Query: 428 RSLSYQVEVLSKLKHPNLITLIGVNQES--KTLIYEYLPNGSLEDHLSRNGNNNAPPLTW 485
R ++ +LS ++H NL+ L+G ES + L+Y ++ NGSL+D L L W
Sbjct: 2280 REFDNELNLLSAIQHENLVPLLGYCNESDQQILVYPFMSNGSLQDRLYGEPAKRKI-LDW 2456
Query: 486 QTRIRIATELCSALIFLHSNKPHSIVHGDLKPSNILLDANLVTKLSDFGICRVLSCQNDS 545
TR+ IA L +LH+ S++H D+K SNILLD ++ K++DFG + + DS
Sbjct: 2457 PTRLSIALGAARGLAYLHTFPGRSVIHRDIKSSNILLDHSMCAKVADFGFSKYAPQEGDS 2636
Query: 546 SSNNSTTQFWITSFAKGTFAYMDPEFLGTGELTSKSDVYSFGIVLLRLITGKPALGIKN- 604
+++ +GT Y+DPE+ T +L+ KSDV+SFG+VLL +++G+ L IK
Sbjct: 2637 ---------YVSLEVRGTAGYLDPEYYKTQQLSEKSDVFSFGVVLLEIVSGREPLNIKRP 2789
Query: 605 EVLYALNN------AGGNVKSVLDP-LAGDWPIVEAEKLVHFALRCCDMNKKSRPEL 654
++L G V ++DP + G + ++V AL+C + RP +
Sbjct: 2790 RTEWSLVEWATPYIRGSKVDEIVDPGIKGGYHAEAMWRVVEVALQCLEPFSTYRPSM 2960
>TC8971 similar to GB|AAM16251.1|20334780|AY093990 At2g11520/F14P14.15
{Arabidopsis thaliana;}, partial (37%)
Length = 1079
Score = 124 bits (311), Expect = 5e-29
Identities = 81/238 (34%), Positives = 129/238 (54%), Gaps = 12/238 (5%)
Frame = +3
Query: 433 QVEVLSKLKHPNLITLIGVNQES--KTLIYEYLPNGSLEDHLSRNGNNNAPPLTWQTRIR 490
+VE+L+K+ H NL+ L+G + + LI EY+PNG+L +HL L + R+
Sbjct: 6 EVELLAKIDHRNLVKLLGFIDKGNERILITEYVPNGTLREHLD---GLRGKILDFNQRLE 176
Query: 491 IATELCSALIFLHSNKPHSIVHGDLKPSNILLDANLVTKLSDFGICRVLSCQNDSSSNNS 550
IA ++ L +LH I+H D+K SNILL ++ K++DFG R+ D
Sbjct: 177 IAIDVAHGLTYLHLYAEKQIIHRDVKSSNILLTESMRAKVADFGFARLGPVNGD------ 338
Query: 551 TTQFWITSFAKGTFAYMDPEFLGTGELTSKSDVYSFGIVLLRLITGKPALGIKN------ 604
Q I++ KGT Y+DPE++ T +LT KSDVYSFGI+LL ++TG+ + +K
Sbjct: 339 --QTHISTKVKGTVGYLDPEYMKTHQLTPKSDVYSFGILLLEILTGRRPVELKKAADERV 512
Query: 605 EVLYALNNAG-GNVKSVLDPLAGDWPIVEAE---KLVHFALRCCDMNKKSRPELCSEG 658
+ +A G+V ++DPL + V A+ K++ + +C + RP + S G
Sbjct: 513 TLRWAFRKYNEGSVVELMDPLMEE--AVNADVLMKMLDLSFQCAAPIRTDRPNMKSVG 680
>TC10873 similar to UP|Q9LDZ5 (Q9LDZ5) F2D10.13 (F5M15.3), partial (77%)
Length = 1478
Score = 124 bits (311), Expect = 5e-29
Identities = 80/231 (34%), Positives = 124/231 (53%), Gaps = 11/231 (4%)
Frame = +1
Query: 433 QVEVLSKLKHPNLITLIG--VNQESKTLIYEYLPNGSLEDHLSRNGNNNAPPLTWQTRIR 490
+V +LS L H NL+ LIG + + + L+YEY+P GSLEDHL ++ P L W TR++
Sbjct: 583 EVLMLSLLHHTNLVRLIGYCTDGDQRLLVYEYMPMGSLEDHLFELSHDKEP-LNWSTRMK 759
Query: 491 IATELCSALIFLHSNKPHSIVHGDLKPSNILLDANLVTKLSDFGICRVLSCQNDSSSNNS 550
+A L +LH +++ DLK +NILLD KLSDFG+ ++ +++
Sbjct: 760 VAVGAARGLEYLHCTADPPVIYRDLKSANILLDNEFNPKLSDFGLAKLGPVGDNTH---- 927
Query: 551 TTQFWITSFAKGTFAYMDPEFLGTGELTSKSDVYSFGIVLLRLITGKPAL------GIKN 604
+++ GT+ Y PE+ +G+LT KSD+YSFG+VLL L+TG+ A+ G +N
Sbjct: 928 -----VSTRVMGTYGYCAPEYAMSGKLTLKSDIYSFGVVLLELLTGRRAIDTSRRPGEQN 1092
Query: 605 EVLYALNNAGGNVK--SVLDP-LAGDWPIVEAEKLVHFALRCCDMNKKSRP 652
V +A + ++DP L G +P + + C K RP
Sbjct: 1093LVSWARPYFSDRRRFGHMVDPLLQGRFPSRCLHQAIAITAMCLQEQPKFRP 1245
>TC16203 UP|Q8GRU6 (Q8GRU6) LRR receptor-like kinase (Hypernodulation aberrant
root formation protein), complete
Length = 3308
Score = 122 bits (305), Expect = 2e-28
Identities = 77/238 (32%), Positives = 127/238 (53%), Gaps = 16/238 (6%)
Frame = +2
Query: 433 QVEVLSKLKHPNLITLIGV--NQESKTLIYEYLPNGSLED--HLSRNGNNNAPPLTWQTR 488
++E L K++H N++ L+G N+++ L+YEY+PNGSL + H ++ G+ L W+ R
Sbjct: 2327 EIETLGKIRHRNIMRLLGYVSNKDTNLLLYEYMPNGSLGEWLHGAKGGH-----LRWEMR 2491
Query: 489 IRIATELCSALIFLHSNKPHSIVHGDLKPSNILLDANLVTKLSDFGICRVLSCQNDSSSN 548
+IA E L ++H + I+H D+K +NILLDA+ ++DFG+ + L S S
Sbjct: 2492 YKIAVEAARGLCYMHHDCSPLIIHRDVKSNNILLDADFEAHVADFGLAKFLYDPGASQSM 2671
Query: 549 NSTTQFWITSFAKGTFAYMDPEFLGTGELTSKSDVYSFGIVLLRLITG-KPA-------- 599
+S G++ Y+ PE+ T ++ KSDVYSFG+VLL LI G KP
Sbjct: 2672 SSIA---------GSYGYIAPEYAYTLKVDEKSDVYSFGVVLLELIIGRKPVGEFGDGVD 2824
Query: 600 -LGIKNEVLYALNNAGGN--VKSVLDPLAGDWPIVEAEKLVHFALRCCDMNKKSRPEL 654
+G N+ + L+ V +V+DP +P+ + + A+ C +RP +
Sbjct: 2825 IVGWVNKTMSELSQPSDTALVLAVVDPRLSGYPLTSVIHMFNIAMMCVKEMGPARPTM 2998
>TC17194 UP|CAE02590 (CAE02590) Nod-factor receptor 1b, complete
Length = 2193
Score = 117 bits (292), Expect = 8e-27
Identities = 78/239 (32%), Positives = 127/239 (52%), Gaps = 15/239 (6%)
Frame = +1
Query: 433 QVEVLSKLKHPNLITLIGVNQE-SKTLIYEYLPNGSLEDHLSRNGNNNAPPLTWQTRIRI 491
+++VL+ + H NL+ LIG E S L+YE++ NG+L +L +G PL W +R++I
Sbjct: 1207 ELKVLTHVHHLNLVRLIGYCVEGSLFLVYEHIDNGNLGQYLHGSGKE---PLPWSSRVQI 1377
Query: 492 ATELCSALIFLHSNKPHSIVHGDLKPSNILLDANLVTKLSDFGICRVLSCQNDSSSNNST 551
A + L ++H + +H D+K +NIL+D NL K++DFG+ +++ NST
Sbjct: 1378 ALDAARGLEYIHEHTVPVYIHRDVKSANILIDKNLRGKVADFGLTKLIEV------GNST 1539
Query: 552 TQFWITSFAKGTFAYMDPEFLGTGELTSKSDVYSFGIVLLRLITGKPALGIKNEVL---- 607
Q + GTF YM PE+ G+++ K DVY+FG+VL LI+ K A+ E++
Sbjct: 1540 LQTRLV----GTFGYMPPEYAQYGDISPKIDVYAFGVVLFELISAKNAVLKTGELVAESK 1707
Query: 608 -------YALNNAG--GNVKSVLDPLAGD-WPIVEAEKLVHFALRCCDMNKKSRPELCS 656
ALN + ++ ++DP G+ +PI K+ C N RP + S
Sbjct: 1708 GLVALFEEALNKSDPCDALRKLVDPRLGENYPIDSVLKIAQLGRACTRDNPLLRPSMRS 1884
>TC14667 homologue to UP|Q84P43 (Q84P43) Protein kinase Pti1, complete
Length = 1591
Score = 115 bits (289), Expect = 2e-26
Identities = 85/250 (34%), Positives = 127/250 (50%), Gaps = 14/250 (5%)
Frame = +2
Query: 419 VLKDPQNFNRSLSYQVEVLSKLKHPNLITLIG--VNQESKTLIYEYLPNGSLEDHL-SRN 475
V +P+ N L+ QV ++S+LK+ N + L G V + L YE+ GSL D L R
Sbjct: 455 VSSEPETNNEFLT-QVSMVSRLKNDNFVELHGYCVEGNLRVLAYEFATMGSLHDILHGRK 631
Query: 476 GNNNAPP---LTWQTRIRIATELCSALIFLHSNKPHSIVHGDLKPSNILLDANLVTKLSD 532
G A P L W R+RIA + L +LH +I+H D++ SN+L+ + K++D
Sbjct: 632 GVQGAQPGPTLDWIQRVRIAVDAARGLEYLHEKVQPAIIHRDIRSSNVLIFEDYKAKIAD 811
Query: 533 FGICRVLSCQNDSSSNNSTTQFWITSFAKGTFAYMDPEFLGTGELTSKSDVYSFGIVLLR 592
F + D ++ +T+ GTF Y PE+ TG+LT KSDVYSFG+VLL
Sbjct: 812 FNLSNQAP---DMAARLHSTR------VLGTFGYHAPEYAMTGQLTQKSDVYSFGVVLLE 964
Query: 593 LITGKPAL------GIKNEVLYALNN-AGGNVKSVLDP-LAGDWPIVEAEKLVHFALRCC 644
L+TG+ + G ++ V +A + VK +DP L G++P KL A C
Sbjct: 965 LLTGRKPVDHTMPRGQQSLVTWATPRLSEDKVKQCVDPKLKGEYPPKGVAKLAAVAALCV 1144
Query: 645 DMNKKSRPEL 654
+ RP +
Sbjct: 1145QYEAEFRPNM 1174
>TC19002 weakly similar to UP|Q9XJ10 (Q9XJ10) ESTs C22458(C62866), partial
(48%)
Length = 780
Score = 105 bits (263), Expect = 2e-23
Identities = 65/212 (30%), Positives = 110/212 (51%), Gaps = 4/212 (1%)
Frame = +1
Query: 433 QVEVLSKLKHPNLITLIGVNQES--KTLIYEYLPNGSLEDHLSRNGNNNAPPLTWQTRIR 490
+VE+L++++H NL++L G E + ++Y+Y+PN SL HL ++ L W R+
Sbjct: 196 EVEILARVRHKNLLSLRGYCAEGQERLIVYDYMPNLSLLSHLHGQHSSECL-LDWNRRMN 372
Query: 491 IATELCSALIFLHSNKPHSIVHGDLKPSNILLDANLVTKLSDFGICRVLSCQNDSSSNNS 550
IA +++LH I+H D+K SN+LLD++ +++DFG +++ D +++
Sbjct: 373 IAIGSAEGIVYLHHQATPHIIHRDIKASNVLLDSDFQARVADFGFAKLIP---DGATH-- 537
Query: 551 TTQFWITSFAKGTFAYMDPEFLGTGELTSKSDVYSFGIVLLRLITGKPALGIKNEVLYAL 610
+T+ KGT Y+ PE+ G+ DV+SFGI+LL L +GK L
Sbjct: 538 -----VTTRVKGTLGYLAPEYAMLGKANECCDVFSFGILLLELASGKKPL---------- 672
Query: 611 NNAGGNVKSVLDPLAGDW--PIVEAEKLVHFA 640
+ S + DW P+ A+K FA
Sbjct: 673 ----EKLSSTVKRSINDWALPLACAKKFTEFA 756
>AV417846
Length = 429
Score = 105 bits (263), Expect = 2e-23
Identities = 60/145 (41%), Positives = 86/145 (58%)
Frame = +1
Query: 462 YLPNGSLEDHLSRNGNNNAPPLTWQTRIRIATELCSALIFLHSNKPHSIVHGDLKPSNIL 521
++P GS+E+HL R G+ P +W R++IA L FLHS +P +++ D K SNIL
Sbjct: 1 FMPKGSMENHLFRRGSY-FQPFSWSLRLKIALGAAKGLAFLHSTEP-KVIYRDFKTSNIL 174
Query: 522 LDANLVTKLSDFGICRVLSCQNDSSSNNSTTQFWITSFAKGTFAYMDPEFLGTGELTSKS 581
LD KLSDFG+ R + S +++ GT Y PE+L TG LT+ S
Sbjct: 175 LDTKYSAKLSDFGLARDGPVGDKSH---------VSTRVMGTRGYAAPEYLATGHLTANS 327
Query: 582 DVYSFGIVLLRLITGKPALGIKNEV 606
DVYSFG+VLL +I+G+ A+ KN++
Sbjct: 328 DVYSFGVVLLEIISGRRAID-KNQL 399
>TC18848 weakly similar to UP|Q9SSQ6 (Q9SSQ6) F6D8.24 protein, partial (64%)
Length = 969
Score = 105 bits (261), Expect = 3e-23
Identities = 58/167 (34%), Positives = 96/167 (56%), Gaps = 2/167 (1%)
Frame = +2
Query: 433 QVEVLSKLKHPNLITLIG--VNQESKTLIYEYLPNGSLEDHLSRNGNNNAPPLTWQTRIR 490
+VEVL +++H NL+ L G V + + ++Y+Y+PN SL HL L WQ R++
Sbjct: 434 EVEVLGRVRHKNLLGLRGYCVGDDQRLIVYDYMPNLSLLSHLHGQFAVEVQ-LNWQKRMK 610
Query: 491 IATELCSALIFLHSNKPHSIVHGDLKPSNILLDANLVTKLSDFGICRVLSCQNDSSSNNS 550
IA +++LH I+H D+K SN+LL+++ ++DFG +++ ++
Sbjct: 611 IAIGSAEGILYLHHEVTPHIIHRDIKASNVLLNSDFEPLVADFGFAKLI----PEGVSHM 778
Query: 551 TTQFWITSFAKGTFAYMDPEFLGTGELTSKSDVYSFGIVLLRLITGK 597
TT+ KGT Y+ PE+ G+++ DVYSFGI+LL L+TG+
Sbjct: 779 TTR------VKGTLGYLAPEYAMWGKVSESCDVYSFGILLLELVTGR 901
>TC9461 similar to UP|O49840 (O49840) Protein kinase (AT2G02800/T20F6.6) ,
partial (37%)
Length = 484
Score = 104 bits (260), Expect = 4e-23
Identities = 57/139 (41%), Positives = 84/139 (60%)
Frame = +3
Query: 462 YLPNGSLEDHLSRNGNNNAPPLTWQTRIRIATELCSALIFLHSNKPHSIVHGDLKPSNIL 521
++P GSLE+HL R G PL+W R+++A L FLH+ K +++ D K SNIL
Sbjct: 3 FMPKGSLENHLFRRGPQ---PLSWSVRMKVAIGAARGLSFLHNAKSQ-VIYRDFKASNIL 170
Query: 522 LDANLVTKLSDFGICRVLSCQNDSSSNNSTTQFWITSFAKGTFAYMDPEFLGTGELTSKS 581
LDA KLSDFG+ + + + + +++ GT Y PE++ TG LT+KS
Sbjct: 171 LDAEFNAKLSDFGLAK---------AGPTGDRTHVSTQVMGTQGYAAPEYVATGRLTAKS 323
Query: 582 DVYSFGIVLLRLITGKPAL 600
DVYSFG+VLL L++G+ A+
Sbjct: 324 DVYSFGVVLLELLSGRRAV 380
>AV419229
Length = 395
Score = 103 bits (257), Expect = 9e-23
Identities = 54/136 (39%), Positives = 81/136 (58%)
Frame = +1
Query: 458 LIYEYLPNGSLEDHLSRNGNNNAPPLTWQTRIRIATELCSALIFLHSNKPHSIVHGDLKP 517
L+YE++PNG+L DHLS + PL++ TR+++A L +LH+ I H D+K
Sbjct: 7 LVYEFMPNGTLRDHLSASSKE---PLSFSTRLKVALGSAKGLAYLHTEADPPIFHRDVKA 177
Query: 518 SNILLDANLVTKLSDFGICRVLSCQNDSSSNNSTTQFWITSFAKGTFAYMDPEFLGTGEL 577
+NILLD+ K++DFG+ R+ + +++ KGT Y+DPE+ T +L
Sbjct: 178 TNILLDSRFSAKVADFGLSRLAPVPD----LEGIVPGHVSTVVKGTPGYLDPEYFLTHKL 345
Query: 578 TSKSDVYSFGIVLLRL 593
T KSDVYS G+VLL L
Sbjct: 346 TDKSDVYSLGVVLLEL 393
>TC17311 similar to UP|Q8K2Y2 (Q8K2Y2) Receptor-interacting serine-threonine
kinase 3, partial (7%)
Length = 606
Score = 103 bits (257), Expect = 9e-23
Identities = 63/176 (35%), Positives = 98/176 (54%), Gaps = 9/176 (5%)
Frame = +3
Query: 433 QVEVLSKLKHPNLITLIGV--NQESKTLIYEYLPNGSLEDHLSR-------NGNNNAPPL 483
+VE+LS ++H N++ L ++ES L+Y+YL N SL+ L + +G+ +
Sbjct: 3 EVEILSNIRHNNIVKLQCCISSEESLLLVYDYLENLSLDRWLHKKSKPPTVSGSVQNNII 182
Query: 484 TWQTRIRIATELCSALIFLHSNKPHSIVHGDLKPSNILLDANLVTKLSDFGICRVLSCQN 543
W R+ IA + L ++H + +VH DLKPSNILLD+ K++DFG+ ++S +
Sbjct: 183 DWPRRLHIAIGVAQGLCYMHHDCSPPVVHRDLKPSNILLDSQFNAKVADFGLA-MMSVKP 359
Query: 544 DSSSNNSTTQFWITSFAKGTFAYMDPEFLGTGELTSKSDVYSFGIVLLRLITGKPA 599
+ + S GTF Y+ PE+ T + K DVYSFG++LL L TGK A
Sbjct: 360 EELATMSA--------VAGTFGYIAPEYALTIRVNEKIDVYSFGVILLELTTGKEA 503
>TC12493 similar to UP|Q9LP77 (Q9LP77) T1N15.9, partial (35%)
Length = 837
Score = 100 bits (249), Expect = 8e-22
Identities = 63/181 (34%), Positives = 90/181 (48%), Gaps = 2/181 (1%)
Frame = +3
Query: 420 LKDPQNFNRSLSYQVEVLSKLKHPNLITLIGV--NQESKTLIYEYLPNGSLEDHLSRNGN 477
LKD ++E + KL H NL+ L G +++ K ++Y+Y+P GSL L N
Sbjct: 288 LKDVTATEMEFREKIEEVGKLVHENLVPLRGYYFSRDEKLVVYDYMPMGSLSALLHANNG 467
Query: 478 NNAPPLTWQTRIRIATELCSALIFLHSNKPHSIVHGDLKPSNILLDANLVTKLSDFGICR 537
PL W+TR IA + +LHS P S HG++K SNILL + ++SD G+
Sbjct: 468 AGRTPLNWETRSAIALGAAHGIAYLHSQGPTS-SHGNIKSSNILLTKSFEPRVSDLGLAY 644
Query: 538 VLSCQNDSSSNNSTTQFWITSFAKGTFAYMDPEFLGTGELTSKSDVYSFGIVLLRLITGK 597
+ TS Y PE +++ K+DVYSFGI+LL L+TGK
Sbjct: 645 LA---------------LPTSTPNRISGYRAPEVTDARKVSQKADVYSFGIMLLELLTGK 779
Query: 598 P 598
P
Sbjct: 780 P 782
>TC9538 similar to UP|RLK5_ARATH (P47735) Receptor-like protein kinase 5
precursor , partial (10%)
Length = 1067
Score = 97.4 bits (241), Expect = 7e-21
Identities = 59/156 (37%), Positives = 83/156 (52%), Gaps = 8/156 (5%)
Frame = +2
Query: 452 NQESKTLIYEYLPNGSLEDHLSRNGNNNAPP--------LTWQTRIRIATELCSALIFLH 503
N+ S L+YEYL N SL+ L +++ L W R++IA L ++H
Sbjct: 35 NEASMLLVYEYLENHSLDKWLHLKPKSSSVSGVVQQYTVLDWPKRLKIAIGAAQGLSYMH 214
Query: 504 SNKPHSIVHGDLKPSNILLDANLVTKLSDFGICRVLSCQNDSSSNNSTTQFWITSFAKGT 563
+ IVH D+K SNILLD K++DFG+ R+L + + I S GT
Sbjct: 215 HDCSPPIVHRDVKTSNILLDKQFNAKVADFGLARMLIKPGELN---------IMSTVIGT 367
Query: 564 FAYMDPEFLGTGELTSKSDVYSFGIVLLRLITGKPA 599
F Y+ PE++ T ++ K DVYSFG+VLL L TGK A
Sbjct: 368 FGYIAPEYVQTTRISEKVDVYSFGVVLLELTTGKEA 475
>TC15149 similar to UP|Q9LVI6 (Q9LVI6) Probable receptor-like protein kinase
protein (AT3g17840/MEB5_6), partial (45%)
Length = 1489
Score = 96.7 bits (239), Expect = 1e-20
Identities = 62/189 (32%), Positives = 96/189 (49%), Gaps = 3/189 (1%)
Frame = +3
Query: 420 LKDPQNFNRSLSYQVEVLSKLKHPNLITLIGV--NQESKTLIYEYLPNGSLEDHLSRNGN 477
LKD + ++E + + H +L+ L +++ K L+Y+Y+P GSL L N
Sbjct: 366 LKDVTISEKEFKDKIETVGAMDHQSLVPLRAYYFSRDEKLLVYDYMPMGSLSALLHGNKG 545
Query: 478 NNAPPLTWQTRIRIATELCSALIFLHSNKPHSIVHGDLKPSNILLDANLVTKLSDFGICR 537
PL W+ R IA + +LHS P ++ HG++K SNILL + K+SDFG+
Sbjct: 546 AGRTPLNWEIRSGIALGAARGIEYLHSQGP-NVSHGNIKASNILLTKSYEAKVSDFGLAH 722
Query: 538 VLSCQNDSSSNNSTTQFWITSFAKGTFAYMDPEFLGTGELTSKSDVYSFGIVLLRLITGK 597
++ SS+ N Y PE +++ K+DVYSFG++LL L+TGK
Sbjct: 723 LV---GPSSTPNRVA------------GYRAPEVTDPRKVSQKADVYSFGVLLLELLTGK 857
Query: 598 -PALGIKNE 605
P + NE
Sbjct: 858 APTHALLNE 884
>TC17497 UP|CAE45595 (CAE45595) S-receptor kinase-like protein 2, complete
Length = 2946
Score = 95.1 bits (235), Expect = 3e-20
Identities = 65/234 (27%), Positives = 114/234 (47%), Gaps = 10/234 (4%)
Frame = +1
Query: 433 QVEVLSKLKHPNLITLIGVNQESKTLIYEYLPNGSLEDHLSRNGNNNAPPLTWQTRIRIA 492
+++++++L+H NL+ L G + + L D + + + W R++I
Sbjct: 1804 EIKLIARLQHRNLVKLFGCSVHQDENSHANKKMKILLD------STRSKLVDWNKRLQII 1965
Query: 493 TELCSALIFLHSNKPHSIVHGDLKPSNILLDANLVTKLSDFGICRVLSCQNDSSSNNSTT 552
+ L++LH + I+H DLK SNILLD + K+SDFG+ R+ +
Sbjct: 1966 DGIARGLLYLHQDSRLRIIHRDLKTSNILLDDEMNPKISDFGLARIFIGDQVEAR----- 2130
Query: 553 QFWITSFAKGTFAYMDPEFLGTGELTSKSDVYSFGIVLLRLITGKPALGIKNEVLYALNN 612
T GT+ YM PE+ G + KSDV+SFG+++L +I+GK +G + + LN
Sbjct: 2131 ----TKRVMGTYGYMPPEYAVHGSFSIKSDVFSFGVIVLEIISGK-KIGRFYDPHHHLNL 2295
Query: 613 AGGNVK--------SVLDPLAGDWPIVEAE--KLVHFALRCCDMNKKSRPELCS 656
+ ++D L D P++ E + +H AL C ++RP++ S
Sbjct: 2296 LSHAWRLWIEERPLELVDELLDD-PVIPTEILRYIHVALLCVQRRPENRPDMLS 2454
>TC16248 similar to UP|BAC87845 (BAC87845) Leucine-rich repeat receptor-like
protein kinase 1, partial (17%)
Length = 866
Score = 94.7 bits (234), Expect = 4e-20
Identities = 70/230 (30%), Positives = 110/230 (47%), Gaps = 8/230 (3%)
Frame = +2
Query: 433 QVEVLSKLKHPNLITLIGVNQESKT--LIYEYLPNGSLEDHLSRNGNNNAPPLTWQTRIR 490
+++ L++++H N+I L G SK L+YE+L GSL+ L N + A W+ R+
Sbjct: 5 EIQTLTEIRHRNVIKLHGFCLHSKFSFLVYEFLEGGSLDQVL--NSDTQAAAFHWEKRVN 178
Query: 491 IATELCSALIFLHSNKPHSIVHGDLKPSNILLDANLVTKLSDFGICRVLSCQNDSSSNNS 550
+ + +AL ++H + I+H D+ N+LLD +SDFG + L
Sbjct: 179 VVKGVANALSYMHHDCSPPIIHRDISSKNVLLDLQYEAHVSDFGTAKFL---------KP 331
Query: 551 TTQFWITSFAKGTFAYMDPEFLGTGELTSKSDVYSFGIVLLRLITGKPALGIKNEVLYAL 610
+ W T+FA GTF Y PE T ++ K DVYSFG+ L +I G + + ++
Sbjct: 332 GSHTW-TAFA-GTFGYAAPELAQTMQVNEKCDVYSFGVFALEIIMGMHPGDLISSLMSPS 505
Query: 611 NNAGGNVKSVLD-----PLAGDWPIVEAEKLV-HFALRCCDMNKKSRPEL 654
N ++D P + PI E L+ AL C N SRP +
Sbjct: 506 TVPMVNDLLLIDILDQRPHQVEKPIDEEVILIASLALACLRKNPHSRPTM 655
>AV418711
Length = 419
Score = 94.4 bits (233), Expect = 6e-20
Identities = 48/134 (35%), Positives = 78/134 (57%)
Frame = +3
Query: 467 SLEDHLSRNGNNNAPPLTWQTRIRIATELCSALIFLHSNKPHSIVHGDLKPSNILLDANL 526
+L+ HL +G P L+W+ R+ I L +LH+ +++H D+K +NILLD NL
Sbjct: 3 TLKSHLYGSG---FPSLSWKERLDICIGSARGLHYLHTGYAKAVIHRDVKSANILLDDNL 173
Query: 527 VTKLSDFGICRVLSCQNDSSSNNSTTQFWITSFAKGTFAYMDPEFLGTGELTSKSDVYSF 586
+ K++DFG+ S + Q +++ KG+F Y+DPE+ +LT KSDVYSF
Sbjct: 174 MAKVADFGL---------SKTGPELDQTHVSTAVKGSFGYLDPEYFRRQQLTEKSDVYSF 326
Query: 587 GIVLLRLITGKPAL 600
G+VL ++ +P +
Sbjct: 327 GVVLFEVLCARPVI 368
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.331 0.141 0.432
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,842,029
Number of Sequences: 28460
Number of extensions: 166314
Number of successful extensions: 1567
Number of sequences better than 10.0: 242
Number of HSP's better than 10.0 without gapping: 1400
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1419
length of query: 688
length of database: 4,897,600
effective HSP length: 97
effective length of query: 591
effective length of database: 2,136,980
effective search space: 1262955180
effective search space used: 1262955180
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.9 bits)
S2: 58 (26.9 bits)
Medicago: description of AC146341.5


