

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146341.1 - phase: 0 /pseudo
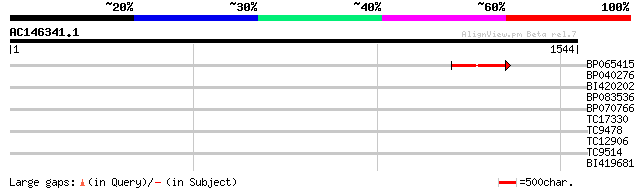
(1544 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BP065415 244 8e-65
BP040276 30 2.9
BI420202 30 2.9
BP083536 30 2.9
BP070766 30 2.9
TC17330 homologue to UP|O65844 (O65844) Protein phosphatase 1, c... 30 3.8
TC9478 similar to UP|R193_ARATH (Q9FNP8) 40S ribosomal protein S... 29 5.0
TC12906 similar to PIR|T00584|T00584 indole-3-acetate beta-gluco... 29 5.0
TC9514 28 8.5
BI419681 28 8.5
>BP065415
Length = 533
Score = 244 bits (623), Expect = 8e-65
Identities = 132/162 (81%), Positives = 140/162 (85%)
Frame = +1
Query: 1203 SNNQSNIMSLNYARCILRMHITCLRLLKEALGERQSRVFDIALATEASNVFAGVFAPSKA 1262
SN S SLNYARCILRMHITCL LLKEALGERQSRVFDIALATEAS AGVFAPSKA
Sbjct: 49 SNIISTTSSLNYARCILRMHITCLCLLKEALGERQSRVFDIALATEASTALAGVFAPSKA 228
Query: 1263 SRAQFQMSSAEVHDTSATNSNELGNNSIKTVVTKATKIAAAVSALVVGAVIYGVTSLERM 1322
SRAQFQMS E HD +AT SN++GNNS K V+ K TKIAAAVSALVVGAVIYGVTSLERM
Sbjct: 229 SRAQFQMSP-EAHDINATISNDVGNNSNKVVLAKTTKIAAAVSALVVGAVIYGVTSLERM 405
Query: 1323 VTILRLKEGLDVIQCIRTTRSNSNGNVRSVGAFKADSSIEVH 1364
VT+LRLKEGLDVIQ +R+TRSNSNGN RSVGAFK D+ IEVH
Sbjct: 406 VTVLRLKEGLDVIQFVRSTRSNSNGNARSVGAFKVDNLIEVH 531
>BP040276
Length = 556
Score = 30.0 bits (66), Expect = 2.9
Identities = 22/83 (26%), Positives = 33/83 (39%)
Frame = +1
Query: 637 NSPSDPRDDANANNAKLPTLKRKRNPASSKDGTSTVSTDQWKTVQSTASSKSAKDGASME 696
NS + NAN K P + N ASS S + D W + +A S + +
Sbjct: 238 NSAPAEETENNANGGKRPAWNKPSNAASS----SVMGADSWPALSKSAHSPA-------K 384
Query: 697 ELKETISVLLQLPNSLSNLNSTG 719
L E++ + + NL TG
Sbjct: 385 SLPESVKGSVDSSSPSPNLQGTG 453
>BI420202
Length = 621
Score = 30.0 bits (66), Expect = 2.9
Identities = 19/57 (33%), Positives = 30/57 (52%), Gaps = 2/57 (3%)
Frame = +1
Query: 860 RDIVFIG--KALKKLRFVEKRAVAAWLLTVVKQVIEETEKNIGKVGQFGRAYSMVDD 914
R++V +G +++KL AW++ KQ++ T KN K GQ G S+V D
Sbjct: 76 REVVLVG**TSMEKL--------VAWIMIANKQIMYSTGKNNAKGGQIGIFGSIVGD 222
>BP083536
Length = 377
Score = 30.0 bits (66), Expect = 2.9
Identities = 17/48 (35%), Positives = 27/48 (55%)
Frame = +3
Query: 198 FRWWYIVRLLQWHHAEGLILPSLVIDWVLNQLQEKDLLEVWELLLPII 245
+R ++ + LL+W+ +ILP LV WVL +L LLLP++
Sbjct: 96 WRRFFPLMLLRWNQMSLMILPMLVFKWVL-------VLMCLRLLLPML 218
>BP070766
Length = 539
Score = 30.0 bits (66), Expect = 2.9
Identities = 13/30 (43%), Positives = 16/30 (53%)
Frame = +2
Query: 828 HMCENKVSCPHHRTPVDGDALRSVDSMRTC 857
HMC N S PH+ T D + RS +S C
Sbjct: 80 HMCINSASFPHNNTSRDEEIARSTESGVNC 169
>TC17330 homologue to UP|O65844 (O65844) Protein phosphatase 1, catalytic
beta subunit (Serine/threonine protein phosphatase) ,
partial (34%)
Length = 499
Score = 29.6 bits (65), Expect = 3.8
Identities = 11/35 (31%), Positives = 21/35 (59%)
Frame = -3
Query: 118 GSVGISSGTADKIQPSRTEIWTKDVIHYLQTLLDE 152
G + ++S + D P +T WT++ IH+ Q L++
Sbjct: 467 GILFLNSASLDPHNPPKTSSWTQEAIHHTQKALED 363
>TC9478 similar to UP|R193_ARATH (Q9FNP8) 40S ribosomal protein S19-3,
partial (97%)
Length = 706
Score = 29.3 bits (64), Expect = 5.0
Identities = 15/40 (37%), Positives = 22/40 (54%), Gaps = 1/40 (2%)
Frame = +2
Query: 102 RATWFVKVTYLNQVRP-GSVGISSGTADKIQPSRTEIWTK 140
RA + VT+ N RP S+ +++ K P+ EIWTK
Sbjct: 344 RAVVLLLVTFFNSCRPCTSLNLTTRVGGKSLPAAVEIWTK 463
>TC12906 similar to PIR|T00584|T00584 indole-3-acetate
beta-glucosyltransferase homolog T27E13.12
- Arabidopsis thaliana {Arabidopsis thaliana;}, partial
(14%)
Length = 675
Score = 29.3 bits (64), Expect = 5.0
Identities = 11/27 (40%), Positives = 16/27 (58%)
Frame = +3
Query: 435 HSVFFLCEWATCDFRDFCTTPPCDIKF 461
H + F W + +FR FC T C++KF
Sbjct: 540 H*IIFRGHWISFEFRTFCCTLLCNMKF 620
>TC9514
Length = 584
Score = 28.5 bits (62), Expect = 8.5
Identities = 22/63 (34%), Positives = 28/63 (43%), Gaps = 1/63 (1%)
Frame = +1
Query: 242 LPIIYGFLEIIVLSQSYVRALAGLTLRVIRDPAPGGSDLVD-NSRRAYTTYALIEMLQYL 300
LP++ L I + S SY+ L G T D GG L S +A+ I LQY
Sbjct: 214 LPVVANTLGIPIKSTSYIPTLNGSTAGKTSDKLKGGGYLQKLFSHKAFDVDTGIRFLQYA 393
Query: 301 ILA 303
LA
Sbjct: 394 GLA 402
>BI419681
Length = 543
Score = 28.5 bits (62), Expect = 8.5
Identities = 21/58 (36%), Positives = 26/58 (44%)
Frame = +1
Query: 671 TVSTDQWKTVQSTASSKSAKDGASMEELKETISVLLQLPNSLSNLNSTGCDESESSVR 728
T ST + K S K EE KE L LP L N NST ++ ESS++
Sbjct: 22 TPSTMEEKHTHIMKDSNYVKSQQEEEEEKEEALSLCDLP--LHNYNSTSLNDDESSLK 189
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.319 0.134 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 25,591,896
Number of Sequences: 28460
Number of extensions: 357677
Number of successful extensions: 1737
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 1718
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1736
length of query: 1544
length of database: 4,897,600
effective HSP length: 102
effective length of query: 1442
effective length of database: 1,994,680
effective search space: 2876328560
effective search space used: 2876328560
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 61 (28.1 bits)
Medicago: description of AC146341.1


