

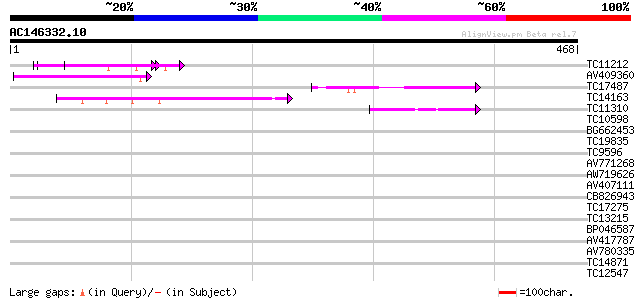
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146332.10 - phase: 0
(468 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC11212 similar to UP|Q84WJ9 (Q84WJ9) At5g19680, partial (38%) 72 2e-13
AV409360 56 1e-08
TC17487 UP|CAE45597 (CAE45597) SAR DNA-binding protein-like prot... 44 7e-05
TC14163 43 1e-04
TC11310 similar to UP|BAD11331 (BAD11331) BRI1-KD interacting pr... 41 5e-04
TC10598 weakly similar to GB|AAM78036.1|21928015|AY125526 At2g01... 39 0.002
BG662453 38 0.003
TC19835 similar to UP|Q9ZRW0 (Q9ZRW0) Nucleolar protein (Fragmen... 37 0.007
TC9596 similar to UP|AAR24722 (AAR24722) At5g49400, partial (66%) 37 0.007
AV771268 37 0.007
AW719626 37 0.007
AV407111 35 0.020
CB826943 35 0.020
TC17275 35 0.026
TC13215 similar to UP|Q39040 (Q39040) Lamin, partial (15%) 34 0.058
BP046587 34 0.058
AV417787 32 0.22
AV780335 32 0.22
TC14871 similar to UP|Q85HQ0 (Q85HQ0) NADH dehydrogenase subunit... 32 0.22
TC12547 32 0.22
>TC11212 similar to UP|Q84WJ9 (Q84WJ9) At5g19680, partial (38%)
Length = 518
Score = 72.0 bits (175), Expect = 2e-13
Identities = 39/104 (37%), Positives = 60/104 (57%), Gaps = 5/104 (4%)
Frame = +1
Query: 46 LDLKFNNLTSLEGLRACVTLKWLSVVENKLESLEGIQGLTKLTVLNAGKNKLKSMDEIGS 105
L L+ N LTS+ GL C+ L+ L + N + +EG+ L L VL+ NKL S+D+I +
Sbjct: 1 LSLQSNRLTSMTGLEGCIALEELYLSHNGITKMEGLSSLVNLRVLDVSSNKLTSVDDIQN 180
Query: 106 LSTIRALILNDNEIVSICNLDQ-----MKELNTLVLSKNPIRKI 144
L+ + L LNDN+I S+ D+ ++L T+ L NP K+
Sbjct: 181 LTQLEDLWLNDNQIDSLEGFDEAVAGSTEKLTTIYLENNPCAKL 312
Score = 51.6 bits (122), Expect = 3e-07
Identities = 34/106 (32%), Positives = 53/106 (49%), Gaps = 5/106 (4%)
Frame = +1
Query: 24 LHLTHKALSDVSCLASFNKLEKLDLKFNNLTSLEGLRACVTLKWLSVVENKLESLEGIQG 83
L L L+ ++ L LE+L L N +T +EGL + V L+ L V NKL S++ IQ
Sbjct: 1 LSLQSNRLTSMTGLEGCIALEELYLSHNGITKMEGLSSLVNLRVLDVSSNKLTSVDDIQN 180
Query: 84 LTKLTVLNAGKNKLKSMDEI-----GSLSTIRALILNDNEIVSICN 124
LT+L L N++ S++ GS + + L +N + N
Sbjct: 181 LTQLEDLWLNDNQIDSLEGFDEAVAGSTEKLTTIYLENNPCAKLSN 318
Score = 41.2 bits (95), Expect = 4e-04
Identities = 29/107 (27%), Positives = 56/107 (52%), Gaps = 5/107 (4%)
Frame = +1
Query: 20 SISSLHLTHKALSDVSCLASFNKLEKLDLKFNNLTSLEGLRACVTLKWLSVVENKLESLE 79
++ L+L+H ++ + L+S L LD+ N LTS++ ++ L+ L + +N+++SLE
Sbjct: 55 ALEELYLSHNGITKMEGLSSLVNLRVLDVSSNKLTSVDDIQNLTQLEDLWLNDNQIDSLE 234
Query: 80 G----IQGLT-KLTVLNAGKNKLKSMDEIGSLSTIRALILNDNEIVS 121
G + G T KLT + N + ++ +R + N +I S
Sbjct: 235 GFDEAVAGSTEKLTTIYLENNPCAKLSNYAAI--LREIFPNIQQIDS 369
>AV409360
Length = 408
Score = 56.2 bits (134), Expect = 1e-08
Identities = 33/119 (27%), Positives = 64/119 (53%), Gaps = 5/119 (4%)
Frame = +1
Query: 4 LSSEQVLKDNNAVNPSSISSLHLTHKALSDVSCLASFNKLEKLDLKFNNLTSL-EGLRAC 62
L+ +++ P + SL+L+ +S + L +L LDL +N + + GL +C
Sbjct: 49 LAGNSIVRITAGALPRGLHSLNLSRNNISSIEGLRELTRLRVLDLSYNRILRIGHGLASC 228
Query: 63 VTLKWLSVVENKLESLEGIQGLTKLTVLNAGKNKLKSMDEIGSL----STIRALILNDN 117
+LK L + NK+ +EG+ L KL++L+ NK+ + +G L ++++A+ L+ N
Sbjct: 229 SSLKELYLAGNKIGEVEGLHRLLKLSILDLRFNKISTAKCLGQLAANYNSLQAINLDGN 405
>TC17487 UP|CAE45597 (CAE45597) SAR DNA-binding protein-like protein, complete
Length = 1794
Score = 43.5 bits (101), Expect = 7e-05
Identities = 40/148 (27%), Positives = 61/148 (41%), Gaps = 9/148 (6%)
Frame = +1
Query: 250 PKLQVFNAKPIDKDTKNEKGHMTDDAHDF--SFDHV-------DQNEDDHLEAADKRKSN 300
PK++ + DKD K G + A + S D V +ED H +ADK+K
Sbjct: 1372 PKIEAY-----DKDRKKGAGGLITPAKTYNPSADAVISQKSDSAMDEDTHEPSADKKKEK 1536
Query: 301 KKRKETADASEKEAGVYDKENTGHNKDNGNKKKDKLTGTVDPDTKNKSTKKKLKKDDNKP 360
K++K+ K N++KD + + K KK KK K
Sbjct: 1537 KEKKK--------------------KKEKNEEKDVPLLADEDGDEEKEVVKKEKKKKRKE 1656
Query: 361 SEKALALEENVNRTEKKKKNRKNKEQSE 388
S + + L+ + EKKKK +K+ E E
Sbjct: 1657 STENVELQNGDDAGEKKKKRKKHAEPEE 1740
Score = 37.7 bits (86), Expect = 0.004
Identities = 28/102 (27%), Positives = 44/102 (42%), Gaps = 6/102 (5%)
Frame = +1
Query: 300 NKKRKETADASEKEAGVYDKENTGHNKDNGNKKKDKLTGTVDPDTKNKSTKKKLKKDDNK 359
+K RK+ A A Y+ + D+ T D K + +KK KK+ N+
Sbjct: 1390 DKDRKKGAGGLITPAKTYNPSADAVISQKSDSAMDEDTHEPSADKKKEKKEKKKKKEKNE 1569
Query: 360 PSEKALALEEN------VNRTEKKKKNRKNKEQSEFDIIDDA 395
+ L +E+ V + EKKKK +++ E E DDA
Sbjct: 1570 EKDVPLLADEDGDEEKEVVKKEKKKKRKESTENVELQNGDDA 1695
>TC14163
Length = 1712
Score = 42.7 bits (99), Expect = 1e-04
Identities = 57/211 (27%), Positives = 90/211 (42%), Gaps = 16/211 (7%)
Frame = +1
Query: 39 SFNKLEKLD-LKFNNLTSLEG-----LRACVTLKWLSVVENKLESL--EGIQGLTKLTVL 90
S +K++ LD NL ++ G L L+++ + N+L E I LT+L VL
Sbjct: 397 SLSKIKNLDGFYLLNLKNISGPFPGFLFKLPKLQFIYIENNQLSGRIPENIGNLTRLDVL 576
Query: 91 NAGKNKLKSM--DEIGSLSTIRALILNDNEIVSI--CNLDQMKELNTLVLSKNPIR-KIG 145
+ N+ +G L+ + L L +N + + ++K L L L N I
Sbjct: 577 SLTGNRFTGTIPSSVGGLTHLTQLQLGNNSLTGTIPATIARLKNLTYLSLEGNQFSGAIP 756
Query: 146 EALKKVKSITKLSLSHCQLEG-IDTSLK-FCVELTELRLAHNDIKS-LPEELMHNSKLRN 202
+ + L LS + G I S+ +L L L HN + +P+ L L
Sbjct: 757 DFFSSFTDLGILRLSRNKFSGKIPASISTLAPKLRYLELGHNQLSGKIPDFLGKFRALDT 936
Query: 203 LDLGNNVIAKWSDIKVLKSLTKLRNLNLQGN 233
LDL +N + K+LTK+ NLNL N
Sbjct: 937 LDLSSNRFSGTVPAS-FKNLTKIFNLNLANN 1026
>TC11310 similar to UP|BAD11331 (BAD11331) BRI1-KD interacting protein 102
(Fragment), partial (13%)
Length = 667
Score = 40.8 bits (94), Expect = 5e-04
Identities = 27/91 (29%), Positives = 43/91 (46%)
Frame = +1
Query: 298 KSNKKRKETADASEKEAGVYDKENTGHNKDNGNKKKDKLTGTVDPDTKNKSTKKKLKKDD 357
K KK+K+ D E D+E K +K K + G+ D D K KKK K D
Sbjct: 82 KKEKKKKKKKDQENGEVASSDEEKAEKEKKKKHKVKVE-DGSPDLDQSEKKKKKK-KDQD 255
Query: 358 NKPSEKALALEENVNRTEKKKKNRKNKEQSE 388
+ + ++ + ++EKK K +KN++ E
Sbjct: 256 AEDNAAEISNGKEDRKSEKKHKKKKNQDAEE 348
Score = 30.8 bits (68), Expect = 0.49
Identities = 27/79 (34%), Positives = 38/79 (47%), Gaps = 11/79 (13%)
Frame = +1
Query: 340 VDPDTKNKSTKKKLKKDDNKPSEKALALEENVNRTEKKKKNRKNKEQSEFDI-------- 391
V+ + + K KKK KK D + E A + EE + EKKKK++ E D+
Sbjct: 64 VEVEGEKKEKKKK-KKKDQENGEVASSDEEKAEK-EKKKKHKVKVEDGSPDLDQSEKKKK 237
Query: 392 ---IDDAEASFAEIFNIKD 407
DAE + AEI N K+
Sbjct: 238 KKKDQDAEDNAAEISNGKE 294
>TC10598 weakly similar to GB|AAM78036.1|21928015|AY125526
At2g01100/F23H14.7 {Arabidopsis thaliana;}, partial
(33%)
Length = 902
Score = 38.9 bits (89), Expect = 0.002
Identities = 34/165 (20%), Positives = 66/165 (39%), Gaps = 13/165 (7%)
Frame = +3
Query: 243 RKIKNALPKLQVFNAKPIDKDTKNEKGHMTDDAH--DFSFDHVDQNEDDHLEAADKRKSN 300
RK+K A K + + D D+ +E + +H H D + + E KRK+
Sbjct: 366 RKLKAAKHKKRADSDSESDYDSDDESKRTSRRSHRKHKKRSHYDSDHEKRKEKLSKRKTK 545
Query: 301 KKRKETADASEKEAGVYDKENTGHNKDNGNKKKDKLTGTVDPDTKNKSTKKKLKKDDNKP 360
K E++D S E+ +E + K +D+++ + D+ + + + +K +K
Sbjct: 546 KWSSESSDFSGDESESRSEEERRRKRKQKKKLRDQVSRSDSSDSDSSADEVSKRKRQHKR 725
Query: 361 SEKALALEENVNRTE-----------KKKKNRKNKEQSEFDIIDD 394
+ + + E K K ++ E SE D+ D
Sbjct: 726 HRVTKSSGSDFSSDEENSPVRKRSHGKHHKRHRHSESSESDLSSD 860
>BG662453
Length = 460
Score = 38.1 bits (87), Expect = 0.003
Identities = 34/131 (25%), Positives = 68/131 (50%), Gaps = 6/131 (4%)
Frame = +2
Query: 84 LTKLTVLNAGKNKLKSM-DEIGSLSTIRALILNDNEIV----SICNLDQMKELNTLVLSK 138
L + VL+ N+L+S+ + +G LS ++ L ++ N I SI N ++ELN +
Sbjct: 35 LLNMVVLDVHSNQLRSLPNSVGCLSKLKVLNVSGNLIEYLPKSIENCRALEELNA---NF 205
Query: 139 NPIRKIGEALK-KVKSITKLSLSHCQLEGIDTSLKFCVELTELRLAHNDIKSLPEELMHN 197
N + ++ + + ++ ++ KLS++ +L + S L L N ++SLP++L +
Sbjct: 206 NKLSQLPDTMGFELLNLKKLSVNSNKLVFLPRSTSHLTSLKILDARLNCLRSLPDDLENL 385
Query: 198 SKLRNLDLGNN 208
L L++ N
Sbjct: 386 INLETLNVSQN 418
>TC19835 similar to UP|Q9ZRW0 (Q9ZRW0) Nucleolar protein (Fragment), partial
(16%)
Length = 550
Score = 37.0 bits (84), Expect = 0.007
Identities = 33/129 (25%), Positives = 53/129 (40%), Gaps = 2/129 (1%)
Frame = +1
Query: 259 PIDKDTKNEKGHM--TDDAHDFSFDHVDQNEDDHLEAADKRKSNKKRKETADASEKEAGV 316
P++ +K K D+ + + D D +E D + KK+KE
Sbjct: 91 PVEASSKKSKKKQKAADEGDEMAVDKPTVTNGDAVE--DHKSEKKKKKEKRK-------- 240
Query: 317 YDKENTGHNKDNGNKKKDKLTGTVDPDTKNKSTKKKLKKDDNKPSEKALALEENVNRTEK 376
D+E N NGN + + KKK +KDDN + +A + ++K
Sbjct: 241 LDQEVAAENGTNGNASEPR-----------SVKKKKDRKDDNGEAVEA------ASESKK 369
Query: 377 KKKNRKNKE 385
KKK++KN E
Sbjct: 370 KKKSKKNAE 396
>TC9596 similar to UP|AAR24722 (AAR24722) At5g49400, partial (66%)
Length = 1196
Score = 37.0 bits (84), Expect = 0.007
Identities = 27/123 (21%), Positives = 53/123 (42%), Gaps = 3/123 (2%)
Frame = +2
Query: 279 SFDHVDQNEDDHLEAADKRKSNKKRKETADA---SEKEAGVYDKENTGHNKDNGNKKKDK 335
S+D + + DD E A KRK +D+ S+ E V++ E+ + +
Sbjct: 557 SYDLENNDVDDKEEKAKSSSKKAKRKHRSDSDSGSDSEDSVFETESGSSSVTGSDYSSGS 736
Query: 336 LTGTVDPDTKNKSTKKKLKKDDNKPSEKALALEENVNRTEKKKKNRKNKEQSEFDIIDDA 395
+ + D++ + +++ KK T+KKKK R++K+ S D+
Sbjct: 737 SSDSSSSDSEEERRRRRKKK------------------TQKKKKGRRHKKYSSSSESSDS 862
Query: 396 EAS 398
E++
Sbjct: 863 ESA 871
>AV771268
Length = 514
Score = 37.0 bits (84), Expect = 0.007
Identities = 21/55 (38%), Positives = 32/55 (58%), Gaps = 5/55 (9%)
Frame = -2
Query: 95 NKLKSMDEIGSLSTIRALILNDNEIVSICNLDQ-----MKELNTLVLSKNPIRKI 144
NKL S+D+I +L+ + L LNDN I S+ D+ ++L T+ L NP K+
Sbjct: 513 NKLTSVDDIQNLTQLEDLWLNDNPIDSLEGFDEAVAGSTEKLTTIYLENNPCAKL 349
>AW719626
Length = 598
Score = 37.0 bits (84), Expect = 0.007
Identities = 17/44 (38%), Positives = 30/44 (67%)
Frame = +3
Query: 13 NNAVNPSSISSLHLTHKALSDVSCLASFNKLEKLDLKFNNLTSL 56
N ++ SS+ SL+L+H +++ L++F+ L+ LDL NNL +L
Sbjct: 423 NGSIPISSLQSLNLSHNRFTNLLHLSAFSNLKSLDLSHNNLGTL 554
>AV407111
Length = 371
Score = 35.4 bits (80), Expect = 0.020
Identities = 38/128 (29%), Positives = 59/128 (45%), Gaps = 5/128 (3%)
Frame = +3
Query: 114 LNDNEIVSICNLDQMKELNTLVLSKNPIRK----IGEALKKVKSITKLSLSHCQLEGIDT 169
L+DNEIV + N+ + L TL+++ N + + IGE L + ++ + L ID
Sbjct: 6 LSDNEIVKLENIPYLNRLGTLLINNNRVTRINPNIGEFLPNLHTLVLTNNRIVNLVEID- 182
Query: 170 SLKFCVELTELRLAHNDIKSLPE-ELMHNSKLRNLDLGNNVIAKWSDIKVLKSLTKLRNL 228
L +L L L N+I P L SKL +L + D K +K+ +L
Sbjct: 183 PLASLPKLQFLSLLDNNITKKPNYRLYVISKLDSL--------RVLDFKKVKNKERLEAK 338
Query: 229 NLQGNPVA 236
NL G+ A
Sbjct: 339 NLFGSEEA 362
>CB826943
Length = 571
Score = 35.4 bits (80), Expect = 0.020
Identities = 35/115 (30%), Positives = 56/115 (48%), Gaps = 6/115 (5%)
Frame = +3
Query: 95 NKLKSMDEIGSLST-IRALILNDNEIVSICNLDQMKELNTLVLSKNPIRK----IGEALK 149
NK+ ++ +G+ L L+DNEIV + N+ + L TL+++ N I + IGE L
Sbjct: 195 NKIAVIENLGATEDQFDTLDLSDNEIVKLENIPYLNRLGTLLINNNRITRINPNIGEFLP 374
Query: 150 KVKSITKLSLSHCQLEGIDTSLKFCVELTELRLAHNDI-KSLPEELMHNSKLRNL 203
+ ++ + L ID L +L L L N+I K L SKL++L
Sbjct: 375 NLHTLVLTNNRIVNLVEID-PLASLPKLQFLSLLDNNITKKANYRLYVISKLKSL 536
>TC17275
Length = 796
Score = 35.0 bits (79), Expect = 0.026
Identities = 44/186 (23%), Positives = 73/186 (38%), Gaps = 16/186 (8%)
Frame = +2
Query: 284 DQNEDDHLEAADKRKSNKKRKETADASEKEAGVYDKENTGHNKDNGNKKKDKLTGTVDPD 343
D + E K+K K T + S K K N N + +K + K T
Sbjct: 170 DTENEQEFEQPLKKKPTKPI--TTNLSSKNQTKSIKSN---NNNLSSKNQTKTTLNASTF 334
Query: 344 TKNKSTKKKLKKDDNKPSEKALALEENVNRTEKKK-----------KNRKNKEQSEFDII 392
K ST K+KK +N S K+ + T KK+ KN+ K + D
Sbjct: 335 KKLNSTSIKIKKLNNSTS-KSFTNSSKTSTTSKKQLDLTKLAGAIAKNKTTKATTATDTK 511
Query: 393 DDAEASFAEIFNIKDQENLNHGGEMKLQDQV-----PKDLKLVSSIETLPVKHKSAKMHN 447
+ + + I ++E LN K Q +D LVS + LP++ + + + +
Sbjct: 512 NKIKVNPGHI----EEEKLNKQSNKKKQSPPNWMFEDEDSDLVSDFKDLPIRFQKSLIPD 679
Query: 448 VESLSS 453
+E +S+
Sbjct: 680 LERIST 697
>TC13215 similar to UP|Q39040 (Q39040) Lamin, partial (15%)
Length = 452
Score = 33.9 bits (76), Expect = 0.058
Identities = 17/48 (35%), Positives = 25/48 (51%)
Frame = -2
Query: 93 GKNKLKSMDEIGSLSTIRALILNDNEIVSICNLDQMKELNTLVLSKNP 140
G+ K MD L I AL + NE+VS C + + L T ++ K+P
Sbjct: 448 GRRSSKRMDNNRDLHPIEALSITTNEVVSPCLIQHYEILPTTIIPKSP 305
>BP046587
Length = 502
Score = 33.9 bits (76), Expect = 0.058
Identities = 17/48 (35%), Positives = 25/48 (51%)
Frame = +3
Query: 93 GKNKLKSMDEIGSLSTIRALILNDNEIVSICNLDQMKELNTLVLSKNP 140
G+ K MD L I AL + NE+VS C + + L T ++ K+P
Sbjct: 243 GRRSSKRMDNNRDLHPIEALSITTNEVVSPCLIQHYEILPTTIIPKSP 386
>AV417787
Length = 310
Score = 32.0 bits (71), Expect = 0.22
Identities = 13/33 (39%), Positives = 21/33 (63%)
Frame = +2
Query: 284 DQNEDDHLEAADKRKSNKKRKETADASEKEAGV 316
D +DD+ + A KR SN+KR+ D S+++ V
Sbjct: 137 DSGDDDYQDIASKRSSNRKRRVVLDFSDEDEDV 235
>AV780335
Length = 398
Score = 32.0 bits (71), Expect = 0.22
Identities = 24/102 (23%), Positives = 52/102 (50%), Gaps = 12/102 (11%)
Frame = +2
Query: 296 KRKSNKKRKETADASEKEAGVYD------KENTGHNKDNG------NKKKDKLTGTVDPD 343
++++ +K KE +KEA + D +EN H K +K K + + +
Sbjct: 41 QKETTRK*KEHDMRVQKEAILEDTIRKLNEENDMHMKKEAISEETISKLKKEYAFHIQKE 220
Query: 344 TKNKSTKKKLKKDDNKPSEKALALEENVNRTEKKKKNRKNKE 385
++ T KKLK+++++ K + LEE + + + +K+++ K+
Sbjct: 221 AISEETIKKLKEENDEHVYKEVTLEE*IKKLQAEKESQTQKQ 346
>TC14871 similar to UP|Q85HQ0 (Q85HQ0) NADH dehydrogenase subunit 4, partial
(4%)
Length = 588
Score = 32.0 bits (71), Expect = 0.22
Identities = 25/72 (34%), Positives = 35/72 (47%), Gaps = 3/72 (4%)
Frame = -2
Query: 299 SNKKRKETADASEKEAGVYDKENTGHNKDNGNKKK---DKLTGTVDPDTKNKSTKKKLKK 355
+NK +KE + G+Y K NK NG KKK K TGT P+ T + K
Sbjct: 530 TNKLQKEFP-----QRGIYIKRQKRKNKKNGKKKKRKERKTTGTPTPNDLKGPTLTRPK- 369
Query: 356 DDNKPSEKALAL 367
D+KP +++L
Sbjct: 368 -DSKPLLTSISL 336
>TC12547
Length = 924
Score = 32.0 bits (71), Expect = 0.22
Identities = 21/75 (28%), Positives = 38/75 (50%)
Frame = -2
Query: 16 VNPSSISSLHLTHKALSDVSCLASFNKLEKLDLKFNNLTSLEGLRACVTLKWLSVVENKL 75
V+PSS S++ +L F K+ ++FN+ TS +GL + +++V
Sbjct: 377 VHPSSAQSINKKFASLKI*KMPRQFTSYRKISIRFNSFTSFQGLGTELFAYTITIV---- 210
Query: 76 ESLEGIQGLTKLTVL 90
ESL ++ LT + +L
Sbjct: 209 ESLSMVRFLTLIELL 165
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.308 0.128 0.345
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,558,822
Number of Sequences: 28460
Number of extensions: 76122
Number of successful extensions: 435
Number of sequences better than 10.0: 73
Number of HSP's better than 10.0 without gapping: 408
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 427
length of query: 468
length of database: 4,897,600
effective HSP length: 94
effective length of query: 374
effective length of database: 2,222,360
effective search space: 831162640
effective search space used: 831162640
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 57 (26.6 bits)
Medicago: description of AC146332.10


