

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC145512.9 - phase: 0
(918 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
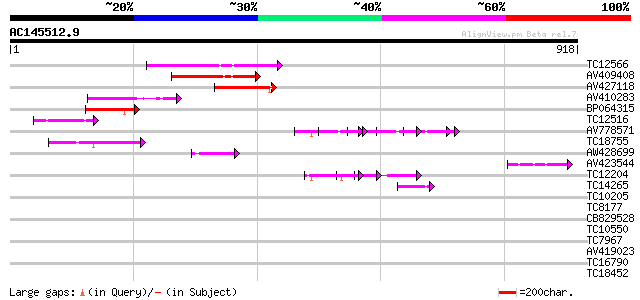
Score E
Sequences producing significant alignments: (bits) Value
TC12566 similar to UP|Q84XI1 (Q84XI1) RCa1 (Fragment), partial (... 124 5e-29
AV409408 124 9e-29
AV427118 89 3e-18
AV410283 86 3e-17
BP064315 74 1e-13
TC12516 similar to UP|Q9M5V7 (Q9M5V7) Disease resistance-like pr... 62 5e-10
AV778571 60 1e-09
TC18755 similar to UP|AAN85378 (AAN85378) Resistance protein (Fr... 58 8e-09
AW428699 57 1e-08
AV423544 49 5e-06
TC12204 weakly similar to UP|Q9ZSN3 (Q9ZSN3) NBS-LRR-like protei... 47 2e-05
TC14265 weakly similar to UP|AAN62760 (AAN62760) Disease resista... 45 4e-05
TC10205 weakly similar to UP|Q8RL77 (Q8RL77) HpaG, partial (10%) 38 0.006
TC8177 homologue to UP|RL15_ARATH (O23515) 60S ribosomal protein... 32 0.35
CB829528 32 0.35
TC10550 weakly similar to UP|Q40392 (Q40392) Virus resistance (N... 32 0.45
TC7967 UP|ATPB_LOTJA (Q9BBU0) ATP synthase beta chain , complete 32 0.59
AV419023 31 0.77
TC16790 31 0.77
TC18452 weakly similar to GB|AAO64875.1|29028992|BT005940 At5g27... 30 1.3
>TC12566 similar to UP|Q84XI1 (Q84XI1) RCa1 (Fragment), partial (28%)
Length = 918
Score = 124 bits (312), Expect = 5e-29
Identities = 84/225 (37%), Positives = 122/225 (53%), Gaps = 5/225 (2%)
Frame = +1
Query: 222 DVLLEQKYGRESDANKLRRLKDLKND---EGYANVLKRVVAYASGHPLALEVIGSHFSNK 278
D + E K D+ +L L K + E Y ++ K++++YA G PLAL+V+G +
Sbjct: 4 DEIYEVKEMSFEDSLQLFSLNAFKRNDPLETYIDLSKKLLSYAKGIPLALKVLGLFLYGR 183
Query: 279 TIEQCNDALDRYKRVPHNMIQTTLQLSYDSLQEEEKIVFLDIACCFKGWKLTMVEGILHA 338
+ L + +++P I L+LSYD L +++K +FLDIAC + G V L +
Sbjct: 184 ERKAWESQLVKLEKLPDPEIINVLKLSYDGLDDDQKDIFLDIACFYLGHLEESVAQTLDS 363
Query: 339 HHGHTMKDQINVLVEKYLIKINESGNVTLHDLVEDMGKEIVRQEAPEDPGKRSRLWFSED 398
G + VL ++ LI I E G + +HDL+E MGKEIVRQ+ P DPGKRSRLW +
Sbjct: 364 C-GFFADIGMEVLKDRCLISILE-GRIVMHDLIEVMGKEIVRQQCPNDPGKRSRLWNDDQ 537
Query: 399 IIQVLEENTGTSKIEMI--HFDGVIEVQWDGEAFKKMENLKTLIF 441
VL +N GT I+ I + + V E FK M NL+ L F
Sbjct: 538 TYDVLGKNKGTDAIQCIFLYTCPIKIVNLHAETFKYMTNLRMLQF 672
>AV409408
Length = 430
Score = 124 bits (310), Expect = 9e-29
Identities = 69/144 (47%), Positives = 93/144 (63%)
Frame = +2
Query: 263 GHPLALEVIGSHFSNKTIEQCNDALDRYKRVPHNMIQTTLQLSYDSLQEEEKIVFLDIAC 322
G PLALEV+GS +++ DAL + K+VPH+ I L++SYD L++E K +FLDIAC
Sbjct: 5 GLPLALEVLGSFLCGRSLSDWEDALIKIKKVPHDDILNKLRISYDMLEDEHKTLFLDIAC 184
Query: 323 CFKGWKLTMVEGILHAHHGHTMKDQINVLVEKYLIKINESGNVTLHDLVEDMGKEIVRQE 382
FKGW V +L + H INVL+EK L+ + + +HDL+E+MGK V E
Sbjct: 185 FFKGWYKHKVIQLLGSCGLHPTVG-INVLIEKSLLTF-DGRVIGMHDLLEEMGKTEVFLE 358
Query: 383 APEDPGKRSRLWFSEDIIQVLEEN 406
+P DPG+RSRLW EDI QVL +N
Sbjct: 359 SPNDPGRRSRLWSLEDIDQVLRKN 430
>AV427118
Length = 317
Score = 89.0 bits (219), Expect = 3e-18
Identities = 50/106 (47%), Positives = 66/106 (62%), Gaps = 6/106 (5%)
Frame = +3
Query: 332 VEGILHAHHGHTMKDQINVLVEKYLIKINESGNVTLHDLVEDMGKEIVRQEAPEDPGKRS 391
V+ IL AH+ + D I+ LV K+LI I+ SG +TLH+ + +MGKEIVR+++ P S
Sbjct: 6 VQHILRAHYRECVTDYISALVSKFLINISSSGELTLHEWMRNMGKEIVRRKSSRMPCVSS 185
Query: 392 RLWFSEDIIQVLEENTGTSKIEMIHFD------GVIEVQWDGEAFK 431
RLW +DI QVLE+ TGT KI I D G I WDG+ FK
Sbjct: 186 RLWILQDIRQVLEDCTGTHKIRTICLDLSSTEEGTI--SWDGKGFK 317
>AV410283
Length = 426
Score = 85.5 bits (210), Expect = 3e-17
Identities = 57/153 (37%), Positives = 79/153 (51%)
Frame = +1
Query: 126 ITSVGEGISILKKRLPEKKVLLLLDDVDKKEQLKAIAGSSNWFRKGSRVIITTRYKNLLI 185
+ V +G+S +K+ L KVLL+LDDVD+ +QL + G+ WF KGSRV+ITTR +L
Sbjct: 40 VNDVNDGVSAIKRVLQGNKVLLILDDVDEIQQLDFLMGNREWFHKGSRVVITTRNTQVLP 219
Query: 186 SSGVERIYEVKGLNDEDAFDLVGWKTLKNDCSPRYKDVLLEQKYGRESDANKLRRLKDLK 245
S V+ YEV+ L A L C + +RR K
Sbjct: 220 ESYVDMFYEVRELELSAALALF--------CH------------------HAMRRKKPA- 318
Query: 246 NDEGYANVLKRVVAYASGHPLALEVIGSHFSNK 278
EG++N+ K++V G PLALEVIGS +K
Sbjct: 319 --EGFSNLSKQIVKKTGGLPLALEVIGSFLFDK 411
>BP064315
Length = 350
Score = 73.6 bits (179), Expect = 1e-13
Identities = 39/90 (43%), Positives = 58/90 (64%), Gaps = 4/90 (4%)
Frame = +1
Query: 124 TEITSVGEGISILKKRLPEKKVLLLLDDVDKKEQLKAIAGSSNWFRKGSRVIITTRYKNL 183
T + S G +K+RL K+VLL+LDDVDK +QL+A+AG +WF GSR+IITTR +
Sbjct: 19 TMMGSTFRGDFEIKRRLSNKRVLLVLDDVDKIKQLQALAGGGDWFGSGSRIIITTRDTTM 198
Query: 184 L----ISSGVERIYEVKGLNDEDAFDLVGW 209
L + + + Y+++ LND D+ +L W
Sbjct: 199 LDKHKMDDVIMKKYKMEELNDHDSLELFCW 288
>TC12516 similar to UP|Q9M5V7 (Q9M5V7) Disease resistance-like protein
(Fragment), partial (36%)
Length = 859
Score = 61.6 bits (148), Expect = 5e-10
Identities = 38/106 (35%), Positives = 62/106 (57%)
Frame = +3
Query: 39 LVGVEHQKQHVTLLLNVGSDDSIHKVGILGIGGIGKTTLALEVYNSIVSQFECSCFLEKV 98
L+GV+ H+ LL S D + +GI G+GGIGKTT+A E++N SQ+E CFL KV
Sbjct: 546 LIGVDKSIAHLESLLCQESKD-VRVIGIWGMGGIGKTTIAEEIFNRRHSQYEGICFLSKV 722
Query: 99 RENSDKNGLIYLQKILLSHIFGEKNTEITSVGEGISILKKRLPEKK 144
++ K+G+ L++ L S + + + +I + +K+R+ K
Sbjct: 723 KDEL*KHGMQSLREKLFSTLLAQ-DVKIDTPNGMADDIKRRIGRMK 857
>AV778571
Length = 452
Score = 60.5 bits (145), Expect = 1e-09
Identities = 43/126 (34%), Positives = 65/126 (51%), Gaps = 14/126 (11%)
Frame = +3
Query: 462 ECRNHM*ELDLSYCINLESFSHVVD----------GFGDKLRTMSVRGCFKLKSIPPLKL 511
EC + DLS NLE S V GF +KLR + + GC K+++ PP+KL
Sbjct: 42 ECESITQIPDLSGLPNLEKLSFVYSENLIKIHESVGFLNKLRILDIEGCCKIRTFPPMKL 221
Query: 512 DSLETMDLSCCFRLESFPLVVDGILGKIKTLNVESCHN--LRSIPPL--KLDSLEKLDIS 567
SLE + LS C LESFP IL K++ + + ++ ++ +P L L +L++
Sbjct: 222 ASLEELYLSHCSSLESFP----EILAKMENITLIGVYHTPIKELPSSIGNLTRLRRLELH 389
Query: 568 YCGSLE 573
CG L+
Sbjct: 390 GCGMLQ 407
Score = 57.0 bits (136), Expect = 1e-08
Identities = 32/80 (40%), Positives = 47/80 (58%), Gaps = 1/80 (1%)
Frame = +3
Query: 500 CFKLKSIPPLK-LDSLETMDLSCCFRLESFPLVVDGILGKIKTLNVESCHNLRSIPPLKL 558
C + IP L L +LE + L V G L K++ L++E C +R+ PP+KL
Sbjct: 45 CESITQIPDLSGLPNLEKLSFVYSENLIKIHESV-GFLNKLRILDIEGCCKIRTFPPMKL 221
Query: 559 DSLEKLDISYCGSLESFPQV 578
SLE+L +S+C SLESFP++
Sbjct: 222 ASLEELYLSHCSSLESFPEI 281
Score = 55.8 bits (133), Expect = 3e-08
Identities = 43/126 (34%), Positives = 67/126 (53%), Gaps = 5/126 (3%)
Frame = +3
Query: 547 CHNLRSIPPLK-LDSLEKLDISYCGSLESFPQVEGRFLGKLKTLNVKSCRIMISIPTLML 605
C ++ IP L L +LEKL Y +L + G FL KL+ L+++ C + + P + L
Sbjct: 45 CESITQIPDLSGLPNLEKLSFVYSENLIKIHESVG-FLNKLRILDIEGCCKIRTFPPMKL 221
Query: 606 SLLEELDLSYCLNLENFPLVVDGFLGKLK--TLSAKSCRNLRSIPSL--KLDLLETLDLS 661
+ LEEL LS+C +LE+FP + L K++ TL ++ +PS L L L+L
Sbjct: 222 ASLEELYLSHCSSLESFPEI----LAKMENITLIGVYHTPIKELPSSIGNLTRLRRLELH 389
Query: 662 NCVSLE 667
C L+
Sbjct: 390 GCGMLQ 407
Score = 53.9 bits (128), Expect = 1e-07
Identities = 44/126 (34%), Positives = 62/126 (48%), Gaps = 5/126 (3%)
Frame = +3
Query: 594 CRIMISIPTLM-LSLLEELDLSYCLNLENFPLVVDGFLGKLKTLSAKSCRNLRSIPSLKL 652
C + IP L L LE+L Y NL V GFL KL+ L + C +R+ P +KL
Sbjct: 45 CESITQIPDLSGLPNLEKLSFVYSENLIKIHESV-GFLNKLRILDIEGCCKIRTFPPMKL 221
Query: 653 DLLETLDLSNCVSLESLPLVVDGFLGKLKTLLVTNCHN--LKSIPPL--KCDALETLDLS 708
LE L LS+C SLES P + L K++ + + ++ +K +P L L+L
Sbjct: 222 ASLEELYLSHCSSLESFPEI----LAKMENITLIGVYHTPIKELPSSIGNLTRLRRLELH 389
Query: 709 CCYSLQ 714
C LQ
Sbjct: 390 GCGMLQ 407
Score = 47.4 bits (111), Expect = 1e-05
Identities = 31/92 (33%), Positives = 48/92 (51%), Gaps = 1/92 (1%)
Frame = +3
Query: 638 AKSCRNLRSIPSLK-LDLLETLDLSNCVSLESLPLVVDGFLGKLKTLLVTNCHNLKSIPP 696
+ C ++ IP L L LE L +L + V GFL KL+ L + C +++ PP
Sbjct: 36 SSECESITQIPDLSGLPNLEKLSFVYSENLIKIHESV-GFLNKLRILDIEGCCKIRTFPP 212
Query: 697 LKCDALETLDLSCCYSLQSFSLVADRLWKLVL 728
+K +LE L LS C SL+SF + ++ + L
Sbjct: 213 MKLASLEELYLSHCSSLESFPEILAKMENITL 308
>TC18755 similar to UP|AAN85378 (AAN85378) Resistance protein (Fragment),
complete
Length = 880
Score = 57.8 bits (138), Expect = 8e-09
Identities = 44/167 (26%), Positives = 84/167 (49%), Gaps = 10/167 (5%)
Frame = +3
Query: 64 VGILGIGGIGKTTLALEVYNS-IVSQFECSCFLEKVRENSDKNGLIYLQKILLSHIFGEK 122
+ + G+GG+GKTTL +VY+ +V + +C V ++ + L+ + L +F E
Sbjct: 267 ISVTGMGGMGKTTLVKQVYDDPVVIKHFRACAWITVSQSCEIGELL---RDLARQLFSEI 437
Query: 123 NTEITSVGEGI------SILKKRLPEKKVLLLLDDVDKKEQLKAIAGSSNWFRKGSRVII 176
+ E + I+K L ++ L++ DDV + +A+ + GSR++I
Sbjct: 438 RRPVPLGLENMRCDRLKMIIKDLLQRRRYLVVFDDVWHVREWEAVKYALPDNNCGSRIMI 617
Query: 177 TTRYKNLLISSGVE---RIYEVKGLNDEDAFDLVGWKTLKNDCSPRY 220
TTR +L +S E ++Y ++ L +++A++L KT D P +
Sbjct: 618 TTRRSDLAFTSSTESKGKVYNLQPLKEDEAWELFCRKTFHGDSCPSH 758
>AW428699
Length = 319
Score = 57.4 bits (137), Expect = 1e-08
Identities = 34/78 (43%), Positives = 45/78 (57%)
Frame = +3
Query: 295 HNMIQTTLQLSYDSLQEEEKIVFLDIACCFKGWKLTMVEGILHAHHGHTMKDQINVLVEK 354
HNM++ + SL+ EK VFLDI C +G+ L V+ IL AH+G + D I VLV+K
Sbjct: 96 HNMVKKR----FYSLKMAEKRVFLDIVCYGEGYHLVDVQHILRAHYGCCVTDYIRVLVDK 263
Query: 355 YLIKINESGNVTLHDLVE 372
LI I+ TLH E
Sbjct: 264 SLIHISPRSEKTLHHWAE 317
>AV423544
Length = 443
Score = 48.5 bits (114), Expect = 5e-06
Identities = 37/110 (33%), Positives = 62/110 (55%), Gaps = 5/110 (4%)
Frame = +1
Query: 807 PAIALCVVSPLTWDGSRRHSIRVIINGNTFIYTDGLKMGTKSPLNMYHLHLFHMKMEN-- 864
P+IAL +VS + + S + IING+ + ++ + H +L+ ++++
Sbjct: 4 PSIALFLVSK-SMESSL--GLNFIINGHEYALAHIFHPVYRT-IRPDHAYLYDLQLQGRK 171
Query: 865 FNDNMEKVLFENMWNHAE---VDFGLPFQKSGIHVSKEKSNMKDIRFTNP 911
F D +++ L EN WNHAE V F F +GIH+ K+KS+++ IRFT P
Sbjct: 172 FKD-IDQALLENEWNHAEIILVSFLSTFIGTGIHIFKQKSSIEHIRFTCP 318
>TC12204 weakly similar to UP|Q9ZSN3 (Q9ZSN3) NBS-LRR-like protein cD7
(Fragment), partial (3%)
Length = 328
Score = 46.6 bits (109), Expect = 2e-05
Identities = 28/78 (35%), Positives = 41/78 (51%), Gaps = 6/78 (7%)
Frame = +1
Query: 530 LVVDGI----LGKIKTLNVESCHNLRSIPP--LKLDSLEKLDISYCGSLESFPQVEGRFL 583
LVV G+ L + +L + +C N S P L+ ++ L + C L+SFPQ + L
Sbjct: 16 LVVTGVQLQYLQSLNSLRICNCPNFESFPEGGLRAPNMTNLHLEKCKKLKSFPQQMNKML 195
Query: 584 GKLKTLNVKSCRIMISIP 601
L TLN+K C + SIP
Sbjct: 196 LSLMTLNIKECPELESIP 249
Score = 46.6 bits (109), Expect = 2e-05
Identities = 34/103 (33%), Positives = 53/103 (51%), Gaps = 7/103 (6%)
Frame = +1
Query: 477 NLESFSHVVDG----FGDKLRTMSVRGCFKLKSIPP--LKLDSLETMDLSCCFRLESFPL 530
NLES VV G + L ++ + C +S P L+ ++ + L C +L+SFP
Sbjct: 4 NLESL--VVTGVQLQYLQSLNSLRICNCPNFESFPEGGLRAPNMTNLHLEKCKKLKSFPQ 177
Query: 531 VVDGILGKIKTLNVESCHNLRSIPPLKL-DSLEKLDISYCGSL 572
++ +L + TLN++ C L SIP DSL L+I +C L
Sbjct: 178 QMNKMLLSLMTLNIKECPELESIPEGGFPDSLNLLEIFHCAKL 306
Score = 41.2 bits (95), Expect = 7e-04
Identities = 34/110 (30%), Positives = 47/110 (41%), Gaps = 1/110 (0%)
Frame = +1
Query: 558 LDSLEKLDISYCGSLESFPQVEGRFLGKLKTLNVKSCRIMISIPTLMLSLLEELDLSYCL 617
L SL L I C + ESFP+ G L+ N+ + L L C
Sbjct: 46 LQSLNSLRICNCPNFESFPE------GGLRAPNMTN-----------------LHLEKCK 156
Query: 618 NLENFPLVVDGFLGKLKTLSAKSCRNLRSIPSLKL-DLLETLDLSNCVSL 666
L++FP ++ L L TL+ K C L SIP D L L++ +C L
Sbjct: 157 KLKSFPQQMNKMLLSLMTLNIKECPELESIPEGGFPDSLNLLEIFHCAKL 306
>TC14265 weakly similar to UP|AAN62760 (AAN62760) Disease resistance
protein-like protein MsR1, partial (5%)
Length = 766
Score = 45.4 bits (106), Expect = 4e-05
Identities = 27/63 (42%), Positives = 38/63 (59%), Gaps = 2/63 (3%)
Frame = +1
Query: 628 GFLGKLKTLSAKSCRNLRSIPSL--KLDLLETLDLSNCVSLESLPLVVDGFLGKLKTLLV 685
G L L+ L SC +L+ +P +L L LD+SNC+SL SLP + G L LK+L +
Sbjct: 10 GKLENLELLRLSSCTDLKGLPDSIGRLSKLRLLDISNCISLPSLPEEI-GNLCNLKSLYM 186
Query: 686 TNC 688
T+C
Sbjct: 187 TSC 195
Score = 38.1 bits (87), Expect = 0.006
Identities = 21/45 (46%), Positives = 30/45 (66%)
Frame = +1
Query: 651 KLDLLETLDLSNCVSLESLPLVVDGFLGKLKTLLVTNCHNLKSIP 695
KL+ LE L LS+C L+ LP + G L KL+ L ++NC +L S+P
Sbjct: 13 KLENLELLRLSSCTDLKGLPDSI-GRLSKLRLLDISNCISLPSLP 144
>TC10205 weakly similar to UP|Q8RL77 (Q8RL77) HpaG, partial (10%)
Length = 586
Score = 38.1 bits (87), Expect = 0.006
Identities = 23/57 (40%), Positives = 35/57 (61%), Gaps = 2/57 (3%)
Frame = +3
Query: 558 LDSLEKLDISYCGSLESFPQVEGRFLGKLKTLNVKSCRIMISIPTLMLSL--LEELD 612
L+ ++ LDIS C SL P+ G L KL+ LNV+ C + +PT ++ L LEE++
Sbjct: 30 LEQVQFLDISDCISLSHLPEDMGE-LKKLEKLNVRGCSRLTELPTSIMDLENLEEVE 197
>TC8177 homologue to UP|RL15_ARATH (O23515) 60S ribosomal protein L15,
complete
Length = 896
Score = 32.3 bits (72), Expect = 0.35
Identities = 27/116 (23%), Positives = 53/116 (45%), Gaps = 13/116 (11%)
Frame = -1
Query: 285 DALDRYKRVPHNMIQTTLQLSYDSLQEEEKIVFLDIACCFKGWKLTMVEGILHAHH---- 340
+AL K++ H+ I+ + +++VFL CC GW +V + AH
Sbjct: 704 NALQNIKKMKHHTIEPAESAV---ATKRKRVVFLP--CCPAGWTCFVV-AMSFAHQTPVM 543
Query: 341 --GHTMKDQINVL-------VEKYLIKINESGNVTLHDLVEDMGKEIVRQEAPEDP 387
++ Q +VL ++ ++I + GN+ +DL+ +G+ ++ EDP
Sbjct: 542 FPSRSLSGQFSVLMDWIAQPIDPWVIPYSSVGNIHQNDLIIFVGRILIHPVRIEDP 375
>CB829528
Length = 188
Score = 32.3 bits (72), Expect = 0.35
Identities = 20/46 (43%), Positives = 28/46 (60%), Gaps = 6/46 (13%)
Frame = +2
Query: 617 LNLENFPLVVD-----GFLGKLKTLSAKSCRNLR-SIPSLKLDLLE 656
L+L+N P +V GFL L++LSAK C L+ +P +KL LE
Sbjct: 50 LSLDNCPSLVTIHESVGFLENLRSLSAKGCTQLKIFVPCIKLTSLE 187
>TC10550 weakly similar to UP|Q40392 (Q40392) Virus resistance (N), partial
(5%)
Length = 670
Score = 32.0 bits (71), Expect = 0.45
Identities = 22/56 (39%), Positives = 29/56 (51%), Gaps = 2/56 (3%)
Frame = +3
Query: 702 LETLDLSC--CYSLQSFSLVADRLWKLVLDDCKELQEIKVIPPCLRMLSAVNCTSL 755
LE LDL SL S + RL L L DC++LQ + +PP + L C+SL
Sbjct: 87 LEILDLEQGRIESLPSCIVNLTRLRYLNLSDCEKLQTLPELPPSVETLLTRGCSSL 254
>TC7967 UP|ATPB_LOTJA (Q9BBU0) ATP synthase beta chain , complete
Length = 2457
Score = 31.6 bits (70), Expect = 0.59
Identities = 18/61 (29%), Positives = 31/61 (50%)
Frame = +3
Query: 63 KVGILGIGGIGKTTLALEVYNSIVSQFECSCFLEKVRENSDKNGLIYLQKILLSHIFGEK 122
K+G+ G G+GKT L +E+ N+I V E + + +Y++ + S + EK
Sbjct: 1038 KIGLFGGAGVGKTVLIMELINNIAKAHGGVSVFGGVGERTREGNDLYME-MKESGVINEK 1214
Query: 123 N 123
N
Sbjct: 1215 N 1217
>AV419023
Length = 410
Score = 31.2 bits (69), Expect = 0.77
Identities = 25/69 (36%), Positives = 36/69 (51%), Gaps = 4/69 (5%)
Frame = +3
Query: 608 LEELDLSYCLNLENFPLVVDGFLGKLKTLSAKSCR--NLRSIPSL--KLDLLETLDLSNC 663
L LDLS+C LE V +G+L L + + N +PS +L L L+LS+C
Sbjct: 3 LVSLDLSFCSILE-----VPNAIGRLWCLERLNLQGNNFVELPSTISRLRRLAYLNLSHC 167
Query: 664 VSLESLPLV 672
+ L LPL+
Sbjct: 168 IKLGDLPLL 194
>TC16790
Length = 584
Score = 31.2 bits (69), Expect = 0.77
Identities = 16/40 (40%), Positives = 25/40 (62%)
Frame = +2
Query: 776 FCLPRVPKIPEWFDHKYEAGLSISFWFRNKFPAIALCVVS 815
F P+ P+IP +K +G ++ FWF+NK P+ CVV+
Sbjct: 44 FPAPKQPEIP--MIYKNSSGSAVVFWFKNKNPS---CVVT 148
>TC18452 weakly similar to GB|AAO64875.1|29028992|BT005940 At5g27920
{Arabidopsis thaliana;}, partial (11%)
Length = 698
Score = 30.4 bits (67), Expect = 1.3
Identities = 23/113 (20%), Positives = 49/113 (43%), Gaps = 4/113 (3%)
Frame = +2
Query: 561 LEKLDISYCGSLESFPQVEGRFLGKLKTLNVKSCRIMISIPTLMLSL----LEELDLSYC 616
LE ++ SYC ++ + LKTL ++ C ++ SI +++ L +D+ C
Sbjct: 302 LEIINTSYCTNITDGSLISLSKCRNLKTLEIRGCLLVTSIGLASIAMNCKQLICVDIKKC 481
Query: 617 LNLENFPLVVDGFLGKLKTLSAKSCRNLRSIPSLKLDLLETLDLSNCVSLESL 669
N+++ ++ + K S ++ + L L + L + L+ L
Sbjct: 482 YNIDDSGMIPLAYFSKSLRQINLSYSSVTDVGLLSLASISCLQSFTILHLQGL 640
Score = 30.4 bits (67), Expect = 1.3
Identities = 27/116 (23%), Positives = 49/116 (41%), Gaps = 4/116 (3%)
Frame = +2
Query: 608 LEELDLSYCLNLENFPLVVDGFLGKLKTLSAKSCRNLRSIP----SLKLDLLETLDLSNC 663
LE ++ SYC N+ + L+ LKTL + C + SI ++ L +D+ C
Sbjct: 302 LEIINTSYCTNITDGSLISLSKCRNLKTLEIRGCLLVTSIGLASIAMNCKQLICVDIKKC 481
Query: 664 VSLESLPLVVDGFLGKLKTLLVTNCHNLKSIPPLKCDALETLDLSCCYSLQSFSLV 719
+++ ++ + K + NL L +SC LQSF+++
Sbjct: 482 YNIDDSGMIPLAYFSKSLRQI-----NLSYSSVTDVGLLSLASISC---LQSFTIL 625
Score = 28.5 bits (62), Expect = 5.0
Identities = 15/66 (22%), Positives = 31/66 (46%), Gaps = 4/66 (6%)
Frame = +2
Query: 470 LDLSYCINLESFSHVVDGFGDKLRTMSVRGCFKLKSIP----PLKLDSLETMDLSCCFRL 525
++ SYC N+ S + L+T+ +RGC + SI + L +D+ C+ +
Sbjct: 311 INTSYCTNITDGSLISLSKCRNLKTLEIRGCLLVTSIGLASIAMNCKQLICVDIKKCYNI 490
Query: 526 ESFPLV 531
+ ++
Sbjct: 491 DDSGMI 508
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.322 0.140 0.416
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,229,920
Number of Sequences: 28460
Number of extensions: 252969
Number of successful extensions: 1291
Number of sequences better than 10.0: 57
Number of HSP's better than 10.0 without gapping: 1236
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1285
length of query: 918
length of database: 4,897,600
effective HSP length: 99
effective length of query: 819
effective length of database: 2,080,060
effective search space: 1703569140
effective search space used: 1703569140
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 60 (27.7 bits)
Medicago: description of AC145512.9


