

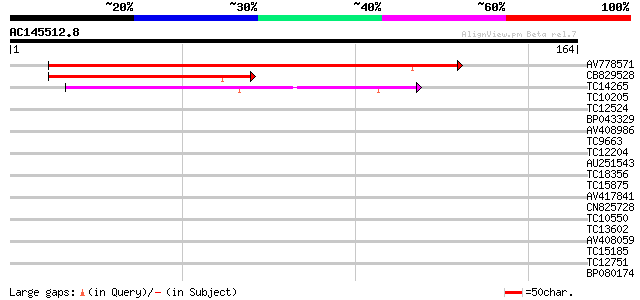
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC145512.8 - phase: 0 /pseudo
(164 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
AV778571 125 4e-30
CB829528 47 2e-06
TC14265 weakly similar to UP|AAN62760 (AAN62760) Disease resista... 45 8e-06
TC10205 weakly similar to UP|Q8RL77 (Q8RL77) HpaG, partial (10%) 34 0.011
TC12524 similar to UP|Q9LKZ6 (Q9LKZ6) Receptor-like protein kina... 34 0.014
BP043329 33 0.019
AV408986 28 1.0
TC9663 similar to UP|Q9FFK7 (Q9FFK7) Signal recognition particle... 28 1.0
TC12204 weakly similar to UP|Q9ZSN3 (Q9ZSN3) NBS-LRR-like protei... 28 1.0
AU251543 27 1.3
TC18356 similar to GB|AAF79264.1|8778255|AC023279 F12K21.25 {Ara... 27 1.3
TC15875 similar to UP|PS21_ARATH (O23714) Proteasome subunit bet... 27 1.3
AV417841 27 1.8
CN825728 27 2.3
TC10550 weakly similar to UP|Q40392 (Q40392) Virus resistance (N... 27 2.3
TC13602 weakly similar to UP|Q84QD8 (Q84QD8) Avr9/Cf-9 rapidly e... 27 2.3
AV408059 26 3.0
TC15185 26 3.9
TC12751 similar to UP|LEU3_BRANA (P29102) 3-isopropylmalate dehy... 25 5.1
BP080174 25 6.7
>AV778571
Length = 452
Score = 125 bits (314), Expect = 4e-30
Identities = 64/121 (52%), Positives = 85/121 (69%), Gaps = 1/121 (0%)
Frame = +3
Query: 12 LTQIPDVSGLVNLEEFSFQGCVNLITVHDSIGLLGRLKTLRVMCCIKLRSIPPLNLASLE 71
+TQIPD+SGL NLE+ SF NLI +H+S+G L +L+ L + C K+R+ PP+ LASLE
Sbjct: 54 ITQIPDLSGLPNLEKLSFVYSENLIKIHESVGFLNKLRILDIEGCCKIRTFPPMKLASLE 233
Query: 72 ELDLSECSCLESFPEILGEMRNVTNLNLEYTPIKELPFPFQNLT-LPKIYPCNCGFVSLP 130
EL LS CS LESFPEIL +M N+T + + +TPIKELP NLT L ++ CG + LP
Sbjct: 234 ELYLSHCSSLESFPEILAKMENITLIGVYHTPIKELPSSIGNLTRLRRLELHGCGMLQLP 413
Query: 131 T 131
+
Sbjct: 414 S 416
>CB829528
Length = 188
Score = 46.6 bits (109), Expect = 2e-06
Identities = 22/61 (36%), Positives = 37/61 (60%), Gaps = 1/61 (1%)
Frame = +2
Query: 12 LTQIPDVSGLVNLEEFSFQGCVNLITVHDSIGLLGRLKTLRVMCCIKLR-SIPPLNLASL 70
LT++P++S L S C +L+T+H+S+G L L++L C +L+ +P + L SL
Sbjct: 5 LTELPNLSAAPFLRNLSLDNCPSLVTIHESVGFLENLRSLSAKGCTQLKIFVPCIKLTSL 184
Query: 71 E 71
E
Sbjct: 185E 187
>TC14265 weakly similar to UP|AAN62760 (AAN62760) Disease resistance
protein-like protein MsR1, partial (5%)
Length = 766
Score = 44.7 bits (104), Expect = 8e-06
Identities = 32/109 (29%), Positives = 54/109 (49%), Gaps = 6/109 (5%)
Frame = +1
Query: 17 DVSGLVNLEEFSFQGCVNLITVHDSIGLLGRLKTLRVMCCIKLRSIPPL--NLASLEELD 74
++ L NLE C +L + DSIG L +L+ L + CI L S+P NL +L+ L
Sbjct: 4 EIGKLENLELLRLSSCTDLKGLPDSIGRLSKLRLLDISNCISLPSLPEEIGNLCNLKSLY 183
Query: 75 LSECSCLESFPEILGEMRNVTNLNLEYTPIK----ELPFPFQNLTLPKI 119
++ C+ E P + ++N+T + E T E P + +P++
Sbjct: 184 MTSCAGCE-LPSSIVNLQNLTVVCDEETAASWEAFEYVIPHLKIEVPQV 327
>TC10205 weakly similar to UP|Q8RL77 (Q8RL77) HpaG, partial (10%)
Length = 586
Score = 34.3 bits (77), Expect = 0.011
Identities = 17/50 (34%), Positives = 29/50 (58%), Gaps = 1/50 (2%)
Frame = +3
Query: 66 NLASLEELDLSECSCLESFPEILGEMRNVTNLNLE-YTPIKELPFPFQNL 114
NL ++ LD+S+C L PE +GE++ + LN+ + + ELP +L
Sbjct: 27 NLEQVQFLDISDCISLSHLPEDMGELKKLEKLNVRGCSRLTELPTSIMDL 176
>TC12524 similar to UP|Q9LKZ6 (Q9LKZ6) Receptor-like protein kinase 1,
partial (27%)
Length = 834
Score = 33.9 bits (76), Expect = 0.014
Identities = 25/86 (29%), Positives = 39/86 (45%), Gaps = 2/86 (2%)
Frame = +2
Query: 16 PDVSGLVNLEEFSFQGCVNLITVHDSIGLLGRLKTLRVMCCIKLRSIPPL--NLASLEEL 73
P++ L L F C + +G L +L TL + + S+ P +L SL+ +
Sbjct: 47 PEIGNLTQLLRFDAAYCGLSGEIPAELGKLQKLDTLFLQVNVLSGSLTPELGHLKSLKSM 226
Query: 74 DLSECSCLESFPEILGEMRNVTNLNL 99
DLS P E++N+T LNL
Sbjct: 227 DLSNNMLSGQVPASFAELKNLTLLNL 304
>BP043329
Length = 484
Score = 33.5 bits (75), Expect = 0.019
Identities = 28/86 (32%), Positives = 39/86 (44%), Gaps = 2/86 (2%)
Frame = +3
Query: 16 PDVSGLVNLEEFSFQGCVNLITVHDSIGLLGRLKTLRVMCCIKLRSIPP--LNLASLEEL 73
P++ L NLE C + + DSIG L +LK L + SIP L SL ++
Sbjct: 87 PEIGNLTNLEVVWLTQCNLVGVIPDSIGNLKKLKDLDLALNDLYGSIPSSLTGLTSLRQI 266
Query: 74 DLSECSCLESFPEILGEMRNVTNLNL 99
+L S P +G N+T L L
Sbjct: 267 ELYNNSLSGELPRGMG---NLTELRL 335
>AV408986
Length = 434
Score = 27.7 bits (60), Expect = 1.0
Identities = 25/81 (30%), Positives = 39/81 (47%), Gaps = 2/81 (2%)
Frame = +3
Query: 21 LVNLEEFSFQGCVNLITVHDSIGLLGRLKTLRVMCCIKLRSIPPL--NLASLEELDLSEC 78
L+ L + SF G + +G L LK L V +IP N + E+DLSE
Sbjct: 51 LLALHQNSFSGAIP-----KELGKLSGLKRLYVYTNQLNGTIPTELGNCTNAIEIDLSEN 215
Query: 79 SCLESFPEILGEMRNVTNLNL 99
+ P+ LG++ N++ L+L
Sbjct: 216 RLIGIIPKELGQISNLSLLHL 278
>TC9663 similar to UP|Q9FFK7 (Q9FFK7) Signal recognition particle receptor
beta subunit-like protein, partial (44%)
Length = 612
Score = 27.7 bits (60), Expect = 1.0
Identities = 12/27 (44%), Positives = 15/27 (55%)
Frame = +1
Query: 25 EEFSFQGCVNLITVHDSIGLLGRLKTL 51
E FSF C N +T D+ GL G + L
Sbjct: 232 EAFSFTQCCNKVTTADASGLTGEISQL 312
>TC12204 weakly similar to UP|Q9ZSN3 (Q9ZSN3) NBS-LRR-like protein cD7
(Fragment), partial (3%)
Length = 328
Score = 27.7 bits (60), Expect = 1.0
Identities = 18/42 (42%), Positives = 24/42 (56%), Gaps = 3/42 (7%)
Frame = +1
Query: 48 LKTLRVMCCIKLRSIPP-LN--LASLEELDLSECSCLESFPE 86
+ L + C KL+S P +N L SL L++ EC LES PE
Sbjct: 127 MTNLHLEKCKKLKSFPQQMNKMLLSLMTLNIKECPELESIPE 252
>AU251543
Length = 360
Score = 27.3 bits (59), Expect = 1.3
Identities = 19/47 (40%), Positives = 26/47 (54%), Gaps = 2/47 (4%)
Frame = -3
Query: 66 NLASLE--ELDLSECSCLESFPEILGEMRNVTNLNLEYTPIKELPFP 110
NL +LE L+LS + L S P+I G + NV + TP+ L FP
Sbjct: 166 NLQALETPSLNLSGLNSLASGPQISGSLCNVGIKVHKQTPLGTLRFP 26
>TC18356 similar to GB|AAF79264.1|8778255|AC023279 F12K21.25 {Arabidopsis
thaliana;} , partial (3%)
Length = 348
Score = 27.3 bits (59), Expect = 1.3
Identities = 17/59 (28%), Positives = 32/59 (53%)
Frame = +1
Query: 11 LLTQIPDVSGLVNLEEFSFQGCVNLITVHDSIGLLGRLKTLRVMCCIKLRSIPPLNLAS 69
+L +I G+++ +EF F+ CV I ++ G ++ + CC KLR + ++L S
Sbjct: 130 MLDKIILTIGVLSFQEFCFRWCVKSIKDK*NV**-GEFCSILLWCCRKLRCVKGISLLS 303
>TC15875 similar to UP|PS21_ARATH (O23714) Proteasome subunit beta type 2-1
(20S proteasome alpha subunit D1) , partial (97%)
Length = 992
Score = 27.3 bits (59), Expect = 1.3
Identities = 14/31 (45%), Positives = 18/31 (57%)
Frame = -1
Query: 109 FPFQNLTLPKIYPCNCGFVSLPTSNYAMSKL 139
FPF + LP I+PCN ++PT Y S L
Sbjct: 929 FPF*QI-LP*IHPCNSLIFTIPTWRYIKSSL 840
>AV417841
Length = 317
Score = 26.9 bits (58), Expect = 1.8
Identities = 12/33 (36%), Positives = 18/33 (54%)
Frame = +1
Query: 76 SECSCLESFPEILGEMRNVTNLNLEYTPIKELP 108
S CL+ L E+R +T N++ P +ELP
Sbjct: 91 SYIECLDGKDASLSEIRRLTGANIQILPREELP 189
>CN825728
Length = 711
Score = 26.6 bits (57), Expect = 2.3
Identities = 15/40 (37%), Positives = 21/40 (52%), Gaps = 4/40 (10%)
Frame = +1
Query: 81 LESFPEILGEMRNVTNLNLEYTPIKELPFPF----QNLTL 116
L P++ R++ +LNL Y K +PF F QNL L
Sbjct: 115 LSPAPDLRLRQRHIHSLNLRYLSSKTMPFQFLWTLQNLYL 234
>TC10550 weakly similar to UP|Q40392 (Q40392) Virus resistance (N), partial
(5%)
Length = 670
Score = 26.6 bits (57), Expect = 2.3
Identities = 12/32 (37%), Positives = 21/32 (65%), Gaps = 2/32 (6%)
Frame = +3
Query: 58 KLRSIPP--LNLASLEELDLSECSCLESFPEI 87
++ S+P +NL L L+LS+C L++ PE+
Sbjct: 114 RIESLPSCIVNLTRLRYLNLSDCEKLQTLPEL 209
>TC13602 weakly similar to UP|Q84QD8 (Q84QD8) Avr9/Cf-9 rapidly elicited
protein 275 (Fragment), partial (26%)
Length = 556
Score = 26.6 bits (57), Expect = 2.3
Identities = 16/38 (42%), Positives = 22/38 (57%), Gaps = 1/38 (2%)
Frame = +3
Query: 66 NLASLEELDLSECSCLES-FPEILGEMRNVTNLNLEYT 102
NLA L+ LDL++ S P +GE +T+LNL T
Sbjct: 348 NLAQLQILDLADNDFNYSQIPSRIGEFSKLTHLNLSLT 461
>AV408059
Length = 280
Score = 26.2 bits (56), Expect = 3.0
Identities = 12/38 (31%), Positives = 20/38 (52%)
Frame = +1
Query: 66 NLASLEELDLSECSCLESFPEILGEMRNVTNLNLEYTP 103
N+ L+ LDLS + +FP L + ++ N+ Y P
Sbjct: 112 NMKCLQNLDLSWNNFSGTFPSSLVNLDELSRFNISYNP 225
>TC15185
Length = 876
Score = 25.8 bits (55), Expect = 3.9
Identities = 13/29 (44%), Positives = 17/29 (57%)
Frame = -2
Query: 28 SFQGCVNLITVHDSIGLLGRLKTLRVMCC 56
S QGC L + SI +L LKT ++CC
Sbjct: 146 SLQGCDILPYLRFSIPILAYLKT*DIICC 60
>TC12751 similar to UP|LEU3_BRANA (P29102) 3-isopropylmalate dehydrogenase,
chloroplast precursor (Beta-IPM dehydrogenase) (IMDH)
(3-IPM-DH) , partial (25%)
Length = 450
Score = 25.4 bits (54), Expect = 5.1
Identities = 12/41 (29%), Positives = 19/41 (46%)
Frame = +2
Query: 99 LEYTPIKELPFPFQNLTLPKIYPCNCGFVSLPTSNYAMSKL 139
L P+K PFP +L P C C + +Y+++ L
Sbjct: 83 LHARPMKASPFPSFSLPRPTSMRCRCSAATPSKKSYSITLL 205
>BP080174
Length = 380
Score = 25.0 bits (53), Expect = 6.7
Identities = 12/29 (41%), Positives = 18/29 (61%)
Frame = +3
Query: 14 QIPDVSGLVNLEEFSFQGCVNLITVHDSI 42
Q P L NL F F+G +N++T+H +I
Sbjct: 276 QTPKSFPLHNL**FLFRGPINIVTLHTTI 362
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.324 0.142 0.430
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,305,915
Number of Sequences: 28460
Number of extensions: 45810
Number of successful extensions: 262
Number of sequences better than 10.0: 45
Number of HSP's better than 10.0 without gapping: 258
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 260
length of query: 164
length of database: 4,897,600
effective HSP length: 83
effective length of query: 81
effective length of database: 2,535,420
effective search space: 205369020
effective search space used: 205369020
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 52 (24.6 bits)
Medicago: description of AC145512.8


