

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC145331.13 + phase: 0 /partial
(365 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
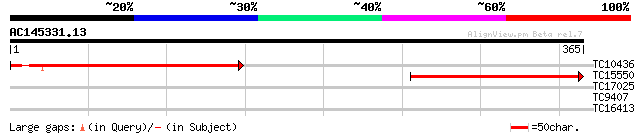
Score E
Sequences producing significant alignments: (bits) Value
TC10436 similar to UP|Q93Z70 (Q93Z70) At2g19940/F6F22.3, partial... 259 5e-70
TC15550 similar to UP|Q93Z70 (Q93Z70) At2g19940/F6F22.3, partial... 202 6e-53
TC17025 similar to UP|Q9ZST2 (Q9ZST2) Pyrophosphate-dependent ph... 31 0.29
TC9407 weakly similar to UP|TAF5_YEAST (P38129) Transcription in... 27 5.4
TC16413 similar to GB|AAF63224.1|7532410|AC002329 3-hydroxy-3-me... 27 7.1
>TC10436 similar to UP|Q93Z70 (Q93Z70) At2g19940/F6F22.3, partial (30%)
Length = 628
Score = 259 bits (662), Expect = 5e-70
Identities = 129/153 (84%), Positives = 139/153 (90%), Gaps = 4/153 (2%)
Frame = +3
Query: 1 DLIKPQNVKLSPIKCSIKS----SQNNVRVGVLGASGYTGSEILRLLANHPQFGVALMTA 56
DL K Q V +KCSIKS SQ VRVGVLGASGYTG+E+LRLLANHP+FGVAL+TA
Sbjct: 180 DLRKQQRV----VKCSIKSGHATSQKTVRVGVLGASGYTGAEVLRLLANHPEFGVALLTA 347
Query: 57 DRKAGQAISSVFPHLGTQDLPDLIAIKDANFSDVDAVFCCLPHGTTQEIIKGLPKHLKIV 116
DRKAGQ+ISSVFPHLG QDLPDLIAIKDANFSDVDAVFCCLPHGTTQ+IIKGLPKHLK+V
Sbjct: 348 DRKAGQSISSVFPHLGPQDLPDLIAIKDANFSDVDAVFCCLPHGTTQDIIKGLPKHLKMV 527
Query: 117 DLSADFRLRDVSEYEEWYGQPHRAPDLQKEAIY 149
DLSADFRLRD+SEYEEWYGQPH+APDLQKEAIY
Sbjct: 528 DLSADFRLRDISEYEEWYGQPHKAPDLQKEAIY 626
>TC15550 similar to UP|Q93Z70 (Q93Z70) At2g19940/F6F22.3, partial (27%)
Length = 617
Score = 202 bits (515), Expect = 6e-53
Identities = 101/110 (91%), Positives = 105/110 (94%)
Frame = +1
Query: 256 IPMSRGMQSTIYVEMAPGVRIDDLRHQLQLSYEDEEFVVLLENGVIPRTHSVKGSNYCLF 315
IPMSRGMQSTIYVEMAPGVRI+DL QL+LSYEDE+FVVLLE GVIPRTHSVKGSNYCL
Sbjct: 1 IPMSRGMQSTIYVEMAPGVRIEDLYEQLKLSYEDEKFVVLLEKGVIPRTHSVKGSNYCLI 180
Query: 316 NVFPDRIPGRAIIISVIDNLVKGASGQALQNLNLIMGFPENLGLQYLPLF 365
NVFPDRIPGRAIIISVIDNLVKGASGQALQNLNLIMGFPE+LGL YLPLF
Sbjct: 181 NVFPDRIPGRAIIISVIDNLVKGASGQALQNLNLIMGFPEDLGLHYLPLF 330
>TC17025 similar to UP|Q9ZST2 (Q9ZST2) Pyrophosphate-dependent
phosphofructokinase alpha subunit , partial (31%)
Length = 649
Score = 31.2 bits (69), Expect = 0.29
Identities = 26/78 (33%), Positives = 30/78 (38%), Gaps = 10/78 (12%)
Frame = +3
Query: 48 QFGVALMTADRKAGQAISSVFPHL----------GTQDLPDLIAIKDANFSDVDAVFCCL 97
+FG A TAD AIS FPH T +PD I + V VFC
Sbjct: 183 EFGDATTTADPSDAPAISRAFPHTYGHPLAHFLRATAKVPDAQIITEHPPMRVGIVFCGR 362
Query: 98 PHGTTQEIIKGLPKHLKI 115
+I GL LKI
Sbjct: 363 QSPGGHNVIWGLHTALKI 416
>TC9407 weakly similar to UP|TAF5_YEAST (P38129) Transcription initiation
factor TFIID subunit 5 (TBP-associated factor 5)
(TBP-associated factor 90 kDa) (TAFII-90), partial (4%)
Length = 530
Score = 26.9 bits (58), Expect = 5.4
Identities = 12/30 (40%), Positives = 18/30 (60%)
Frame = +3
Query: 165 VANPGCYPTSIQLPLVPLIKANLIQTTNII 194
V++ PT +QLPL+PL+ L Q I+
Sbjct: 159 VSSQWTVPTLLQLPLIPLLGFGLCQLVKIL 248
>TC16413 similar to GB|AAF63224.1|7532410|AC002329
3-hydroxy-3-methylglutaryl-coenzyme A reductase 2
{Arabidopsis thaliana;}, partial (41%)
Length = 674
Score = 26.6 bits (57), Expect = 7.1
Identities = 18/59 (30%), Positives = 29/59 (48%)
Frame = +3
Query: 13 IKCSIKSSQNNVRVGVLGASGYTGSEILRLLANHPQFGVALMTADRKAGQAISSVFPHL 71
++ SI S+ ++ +G L SG+T I A+H F +T+ K I+ V P L
Sbjct: 264 VRASISSAARSMALGTLSHSGWTRKFIRVRFASH-TFPAKTLTSSAKTLFGIALVSPPL 437
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.319 0.138 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,535,853
Number of Sequences: 28460
Number of extensions: 64756
Number of successful extensions: 259
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 258
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 259
length of query: 365
length of database: 4,897,600
effective HSP length: 91
effective length of query: 274
effective length of database: 2,307,740
effective search space: 632320760
effective search space used: 632320760
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 56 (26.2 bits)
Medicago: description of AC145331.13


