

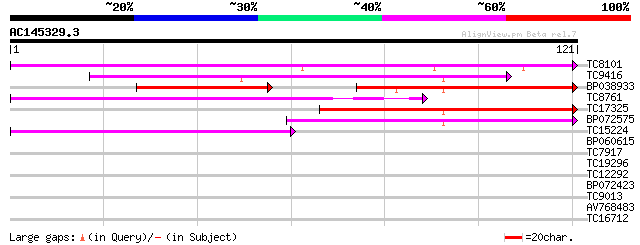
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC145329.3 + phase: 0 /partial
(121 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC8101 similar to UP|O24074 (O24074) DNAJ-like protein, partial ... 64 5e-12
TC9416 similar to UP|Q9FEW7 (Q9FEW7) DnaJ like protein, partial ... 57 7e-10
BP038933 39 2e-08
TC8761 homologue to UP|Q9SWB5 (Q9SWB5) Seed maturation protein P... 47 1e-06
TC17325 similar to UP|Q9M034 (Q9M034) Heat shock protein 40-like... 46 2e-06
BP072575 42 2e-05
TC15224 homologue to UP|Q9SWB5 (Q9SWB5) Seed maturation protein ... 42 2e-05
BP060615 33 0.017
TC7917 similar to UP|Q9M034 (Q9M034) Heat shock protein 40-like,... 32 0.039
TC19296 similar to UP|O22855 (O22855) At2g43410 protein, partial... 27 0.95
TC12292 26 1.6
BP072423 25 2.8
TC9013 24 6.2
AV768483 24 8.1
TC16712 UP|ATPI_LOTJA (Q9BBS5) Chloroplast ATP synthase a chain ... 24 8.1
>TC8101 similar to UP|O24074 (O24074) DNAJ-like protein, partial (90%)
Length = 1737
Score = 64.3 bits (155), Expect = 5e-12
Identities = 41/126 (32%), Positives = 67/126 (52%), Gaps = 5/126 (3%)
Frame = +1
Query: 1 EVLFYEDGEPIIDGESGDLRFRIRTAPHELFKREGNDLHTTVTITLVQALVGFEKTIKHL 60
+++F + D +GD+ F ++ H FKRE +DL+ ++L +AL GF+ +KHL
Sbjct: 850 KIVFEGQADEAPDTITGDIVFVVQVKEHPKFKRELDDLYIDHNLSLTEALCGFQFAVKHL 1029
Query: 61 D--EHLVDISSKGITNPKQVRKFKGEGMPLH-TSTKKGDLYVTFEVLFPTT--LSEEQKT 115
D + L+ + + P Q + EGMP H KG LY+ F V FP + +S +Q
Sbjct: 1030DGRQLLIKSNPGEVIKPGQHKAINDEGMPQHGRPFIKGRLYIKFNVDFPDSGFISPDQCQ 1209
Query: 116 KIKSIL 121
++ IL
Sbjct: 1210LLEKIL 1227
>TC9416 similar to UP|Q9FEW7 (Q9FEW7) DnaJ like protein, partial (26%)
Length = 558
Score = 57.4 bits (137), Expect = 7e-10
Identities = 33/93 (35%), Positives = 50/93 (53%), Gaps = 3/93 (3%)
Frame = +2
Query: 18 DLRFRIRTAPHELFKREGNDLHTTVTITLVQ--ALVGFEKTIKHLDEHLVDISSKGITNP 75
D+ F PH +F R+GNDL T I+L + AL GF + LD + I+ T+P
Sbjct: 2 DIVFIFDEKPHNVFTRDGNDLVVTQKISLTEDEALTGFTFPLTTLDGRCLPIAINNGTDP 181
Query: 76 KQVRKFKGEGMPLHTS-TKKGDLYVTFEVLFPT 107
+ GEGMP+ +K+G+L + F + FP+
Sbjct: 182 NYEKVITGEGMPISKDPSKRGNLRIKFNIKFPS 280
>BP038933
Length = 451
Score = 38.5 bits (88), Expect(2) = 2e-08
Identities = 21/52 (40%), Positives = 32/52 (61%), Gaps = 5/52 (9%)
Frame = -3
Query: 75 PKQVRKF----KGEGMPLHTS-TKKGDLYVTFEVLFPTTLSEEQKTKIKSIL 121
P VR + KGE MP+ +KKG+L + F + FP+ L+ EQK+ +K +L
Sbjct: 305 PSSVRLYEEVVKGEVMPIPKEPSKKGNLRIKFNIKFPSRLTSEQKSGVKRLL 150
Score = 33.5 bits (75), Expect(2) = 2e-08
Identities = 15/29 (51%), Positives = 20/29 (68%)
Frame = -1
Query: 28 HELFKREGNDLHTTVTITLVQALVGFEKT 56
H +FKR+ NDL T I+LV+AL G+ T
Sbjct: 436 HSVFKRDANDLIFTQKISLVEALTGYTAT 350
>TC8761 homologue to UP|Q9SWB5 (Q9SWB5) Seed maturation protein PM37,
partial (74%)
Length = 1050
Score = 46.6 bits (109), Expect = 1e-06
Identities = 29/89 (32%), Positives = 46/89 (51%)
Frame = +3
Query: 1 EVLFYEDGEPIIDGESGDLRFRIRTAPHELFKREGNDLHTTVTITLVQALVGFEKTIKHL 60
++ F + + D +GD+ F ++ H FKR+ DL T++L +AL GF+ + HL
Sbjct: 789 KITFPGEADEAPDTTTGDIVFVLQLKEHPKFKRKAEDLFVEHTLSLTEALCGFQFVLTHL 968
Query: 61 DEHLVDISSKGITNPKQVRKFKGEGMPLH 89
D + I S NP +V K +PLH
Sbjct: 969 DGRQLLIKS----NPGEVVK-----LPLH 1028
>TC17325 similar to UP|Q9M034 (Q9M034) Heat shock protein 40-like, partial
(18%)
Length = 472
Score = 45.8 bits (107), Expect = 2e-06
Identities = 23/56 (41%), Positives = 34/56 (60%), Gaps = 1/56 (1%)
Frame = +1
Query: 67 ISSKGITNPKQVRKFKGEGMPLHTS-TKKGDLYVTFEVLFPTTLSEEQKTKIKSIL 121
+S I +P KGEGMP+ +K+G+L + F + FP+ L+ EQKT IK +L
Sbjct: 16 VSVNNIISPTYEEVIKGEGMPIPKEPSKRGNLRIKFNIKFPSRLTSEQKTGIKRLL 183
>BP072575
Length = 487
Score = 42.4 bits (98), Expect = 2e-05
Identities = 24/63 (38%), Positives = 34/63 (53%), Gaps = 1/63 (1%)
Frame = -2
Query: 60 LDEHLVDISSKGITNPKQVRKFKGEGMPLHTS-TKKGDLYVTFEVLFPTTLSEEQKTKIK 118
LD + I I +P EGMPL +KKG+L + F + FPT L++EQK I+
Sbjct: 486 LDGRNLSILINNIIHPHYEEVVPKEGMPLSKDPSKKGNLRIKFNIKFPTQLNDEQKAGIR 307
Query: 119 SIL 121
+L
Sbjct: 306 KLL 298
>TC15224 homologue to UP|Q9SWB5 (Q9SWB5) Seed maturation protein PM37,
partial (71%)
Length = 1031
Score = 42.4 bits (98), Expect = 2e-05
Identities = 20/61 (32%), Positives = 34/61 (54%)
Frame = +3
Query: 1 EVLFYEDGEPIIDGESGDLRFRIRTAPHELFKREGNDLHTTVTITLVQALVGFEKTIKHL 60
++ F + + D +GD+ F ++ H FKR+ DL T++L +AL GF+ + HL
Sbjct: 843 KITFPGEADEAPDTVTGDIVFVLQQKEHPKFKRKSEDLFVEHTLSLTEALCGFQFVLTHL 1022
Query: 61 D 61
D
Sbjct: 1023D 1025
>BP060615
Length = 454
Score = 32.7 bits (73), Expect = 0.017
Identities = 14/39 (35%), Positives = 26/39 (65%), Gaps = 1/39 (2%)
Frame = -3
Query: 84 EGMPLHTST-KKGDLYVTFEVLFPTTLSEEQKTKIKSIL 121
EGMP+ KKG+L + ++ +P+ L+ EQK+ ++ +L
Sbjct: 440 EGMPISKEPGKKGNLKIKLDIKYPSRLTAEQKSDLRRVL 324
>TC7917 similar to UP|Q9M034 (Q9M034) Heat shock protein 40-like, partial
(10%)
Length = 385
Score = 31.6 bits (70), Expect = 0.039
Identities = 12/30 (40%), Positives = 22/30 (73%)
Frame = -2
Query: 92 TKKGDLYVTFEVLFPTTLSEEQKTKIKSIL 121
++KG+L + F + FP+ L+ EQK+ +K +L
Sbjct: 237 SQKGNLRIKFNIKFPSRLTSEQKSGVKRLL 148
>TC19296 similar to UP|O22855 (O22855) At2g43410 protein, partial (6%)
Length = 481
Score = 26.9 bits (58), Expect = 0.95
Identities = 11/25 (44%), Positives = 20/25 (80%)
Frame = -2
Query: 57 IKHLDEHLVDISSKGITNPKQVRKF 81
+KHL LV+++S+GITNP+ + ++
Sbjct: 267 MKHL---LVNLNSRGITNPQNITRY 202
>TC12292
Length = 587
Score = 26.2 bits (56), Expect = 1.6
Identities = 13/49 (26%), Positives = 24/49 (48%)
Frame = +1
Query: 52 GFEKTIKHLDEHLVDISSKGITNPKQVRKFKGEGMPLHTSTKKGDLYVT 100
G ++ K + H +I G+ Q+ + + + L S +GDLYV+
Sbjct: 361 GIQRRWKEVLHHATEIEEMGVLIQYQLMLMRVQIISLFISNLRGDLYVS 507
>BP072423
Length = 266
Score = 25.4 bits (54), Expect = 2.8
Identities = 11/25 (44%), Positives = 17/25 (68%)
Frame = +1
Query: 62 EHLVDISSKGITNPKQVRKFKGEGM 86
+HL+D +KG T P+Q R+ KG +
Sbjct: 169 QHLIDTGTKGRTKPQQGRR-KGSNL 240
>TC9013
Length = 766
Score = 24.3 bits (51), Expect = 6.2
Identities = 14/35 (40%), Positives = 19/35 (54%)
Frame = -1
Query: 84 EGMPLHTSTKKGDLYVTFEVLFPTTLSEEQKTKIK 118
E PLHT+ + + T EVLF LS + +IK
Sbjct: 724 EKHPLHTTVRNTSV*TTLEVLF-ELLSSPSQMRIK 623
>AV768483
Length = 405
Score = 23.9 bits (50), Expect = 8.1
Identities = 10/15 (66%), Positives = 11/15 (72%)
Frame = +2
Query: 39 HTTVTITLVQALVGF 53
HTT+ TLVQ L GF
Sbjct: 227 HTTINRTLVQKLCGF 271
>TC16712 UP|ATPI_LOTJA (Q9BBS5) Chloroplast ATP synthase a chain precursor
(ATPase subunit IV) , complete
Length = 851
Score = 23.9 bits (50), Expect = 8.1
Identities = 11/35 (31%), Positives = 19/35 (53%)
Frame = +1
Query: 19 LRFRIRTAPHELFKREGNDLHTTVTITLVQALVGF 53
L ++I PH ND++TTV + L+ ++ F
Sbjct: 343 LPWKIIQLPHGELAAPTNDIYTTVALALLTSVAYF 447
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.317 0.138 0.386
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,687,210
Number of Sequences: 28460
Number of extensions: 18159
Number of successful extensions: 83
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 81
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 81
length of query: 121
length of database: 4,897,600
effective HSP length: 79
effective length of query: 42
effective length of database: 2,649,260
effective search space: 111268920
effective search space used: 111268920
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 49 (23.5 bits)
Medicago: description of AC145329.3


