

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC145329.11 - phase: 0
(360 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
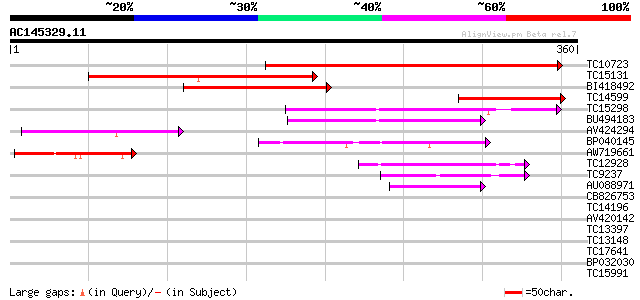
Score E
Sequences producing significant alignments: (bits) Value
TC10723 similar to GB|AAB61117.1|2194142|F20P5 ESTs gb|N38288,gb... 310 3e-85
TC15131 similar to UP|Q9FX71 (Q9FX71) T6J4.1 protein, partial (40%) 171 1e-43
BI418492 149 5e-37
TC14599 weakly similar to UP|Q9P2S7 (Q9P2S7) Cisplatin resistanc... 129 6e-31
TC15298 homologue to UP|QUA1_ARATH (Q9LSG3) Glycosyltransferase ... 88 2e-18
BU494183 87 6e-18
AV424294 81 2e-16
BP040145 80 4e-16
AW719661 64 5e-11
TC12928 49 1e-06
TC9237 weakly similar to UP|GLTR_ARATH (Q9FWA4) Probable glycosy... 46 8e-06
AU088971 44 5e-05
CB826753 27 4.1
TC14196 homologue to UP|Q9FNV7 (Q9FNV7) Auxin-repressed protein,... 27 4.1
AV420142 27 5.3
TC13397 27 5.3
TC13148 similar to UP|Q8LK24 (Q8LK24) SOS2-like protein kinase, ... 27 5.3
TC17641 homologue to GB|AAP31966.1|30387601|BT006622 At3g19640 {... 27 5.3
BP032030 27 6.9
TC15991 similar to GB|CAB10400.1|2244979|ATFCA5 enoyl-CoA hydrat... 27 6.9
>TC10723 similar to GB|AAB61117.1|2194142|F20P5 ESTs
gb|N38288,gb|T43486,gb|AA395242 come from this gene.
{Arabidopsis thaliana;}, partial (49%)
Length = 1008
Score = 310 bits (793), Expect = 3e-85
Identities = 138/190 (72%), Positives = 157/190 (82%), Gaps = 1/190 (0%)
Frame = +1
Query: 163 RNYLADLLESCVKRVIYLDSDLVLQDDIAKLWNTDLGLN-TIGAPQYCHANFTKYFTAAF 221
RNYL D+L+ CV RVIYLDSDLV+ DD++KLWN L + IGAP+YCHANFTKYFT F
Sbjct: 1 RNYLGDMLDPCVDRVIYLDSDLVVVDDVSKLWNVTLAESRVIGAPEYCHANFTKYFTDEF 180
Query: 222 WSDPVFSTTFEKRKACYFNTGVMVMDLVKWRKKGYTERIERWMEIQKVERIYELGSLPPF 281
W DPV S F R+ CYFNTGVMVMDLV+WR+ Y +RIE WME+Q+ +RIYELGSLPPF
Sbjct: 181 WVDPVLSRVFSSRRPCYFNTGVMVMDLVRWREGNYRKRIENWMELQRKKRIYELGSLPPF 360
Query: 282 LLVFAGHVAAIEHRWNQHGLGGDNVKGSCRDLHPGPVSLLHWSGSGKPWRRLDESKPCPL 341
LLVFAG+V AI+HRWNQHGLGGDN+ G CR LHPGPVSLLHWSG GKPW RLD +PCPL
Sbjct: 361 LLVFAGNVEAIDHRWNQHGLGGDNLNGVCRSLHPGPVSLLHWSGKGKPWVRLDNKQPCPL 540
Query: 342 DALWEPFDLY 351
D LWEP+DLY
Sbjct: 541 DRLWEPYDLY 570
>TC15131 similar to UP|Q9FX71 (Q9FX71) T6J4.1 protein, partial (40%)
Length = 553
Score = 171 bits (434), Expect = 1e-43
Identities = 80/149 (53%), Positives = 110/149 (73%), Gaps = 4/149 (2%)
Frame = +1
Query: 51 FHTSPFFRNADQCEPISREIGACH-PSLVHVAITLDVEYLRGSIAAVHSILYHASCPENV 109
F +P FRN +C + +I H PSL+H+A+TLD YLRGS+A V S+L HASCPENV
Sbjct: 88 FREAPAFRNGRECPRTTADIKQAHDPSLIHIAMTLDATYLRGSVAGVFSVLQHASCPENV 267
Query: 110 FFHFLVTDT---DLETLVRTTFPQLRFKVYYFDRNIVKNLISTSVRQALEQPLNYARNYL 166
FHF+ T +L ++ +TFP L F++Y+FD N+V+ IS S+R+AL+QPLNYAR YL
Sbjct: 268 VFHFVATSNRRHELRRVIASTFPYLNFRIYHFDSNLVRGKISFSIRRALDQPLNYARMYL 447
Query: 167 ADLLESCVKRVIYLDSDLVLQDDIAKLWN 195
ADL+ + V+R+IY DSDL++ DD+AKLW+
Sbjct: 448 ADLVPATVRRIIYFDSDLIVVDDVAKLWS 534
>BI418492
Length = 537
Score = 149 bits (377), Expect = 5e-37
Identities = 71/94 (75%), Positives = 87/94 (92%)
Frame = +3
Query: 111 FHFLVTDTDLETLVRTTFPQLRFKVYYFDRNIVKNLISTSVRQALEQPLNYARNYLADLL 170
FHF+V++T+LE+LV++TFP+L+FKVYYF+ + V++LISTSVRQALEQPLNYARNYLADLL
Sbjct: 255 FHFIVSETNLESLVKSTFPELKFKVYYFNPDTVRSLISTSVRQALEQPLNYARNYLADLL 434
Query: 171 ESCVKRVIYLDSDLVLQDDIAKLWNTDLGLNTIG 204
ESCV RVIYLDSDLV+ DDI+KLW+T LG TIG
Sbjct: 435 ESCVGRVIYLDSDLVVVDDISKLWSTSLGSKTIG 536
>TC14599 weakly similar to UP|Q9P2S7 (Q9P2S7) Cisplatin
resistance-associated overexpressed protein, partial
(4%)
Length = 711
Score = 129 bits (325), Expect = 6e-31
Identities = 51/68 (75%), Positives = 59/68 (86%)
Frame = +1
Query: 286 AGHVAAIEHRWNQHGLGGDNVKGSCRDLHPGPVSLLHWSGSGKPWRRLDESKPCPLDALW 345
AG V ++EHRWNQHGLGGDN++G CRDLHPGPVSLLHWSG GKPW R+D KPCPLD+LW
Sbjct: 16 AGEVVSVEHRWNQHGLGGDNLEGLCRDLHPGPVSLLHWSGKGKPWLRIDSKKPCPLDSLW 195
Query: 346 EPFDLYGH 353
P+DL+ H
Sbjct: 196 APYDLFRH 219
>TC15298 homologue to UP|QUA1_ARATH (Q9LSG3) Glycosyltransferase QUASIMODO1
, partial (33%)
Length = 818
Score = 88.2 bits (217), Expect = 2e-18
Identities = 49/181 (27%), Positives = 91/181 (50%), Gaps = 6/181 (3%)
Frame = +2
Query: 176 RVIYLDSDLVLQDDIAKLWNTDLGLNTIGAPQYCHANFTKYFTAAFWSDPVFSTTFEKRK 235
++++LD D+V+Q D+ LW D+ GA + C +F +Y +S P+ F K
Sbjct: 14 KILFLDDDIVVQKDLTGLWKIDMDGKVNGAVETCFGSFHRYAQYMNFSHPLIKAKFNP-K 190
Query: 236 ACYFNTGVMVMDLVKWRKKGYTERIERWMEIQKVERIYELGSLPPFLLVFAGHVAAIEHR 295
AC + G+ DL WR++ TE W + + +++LG+LPP L+ + ++
Sbjct: 191 ACAWAYGMNFFDLDAWRREKCTEEYHYWQNLNENRTLWKLGTLPPGLITYYATTKPLDKS 370
Query: 296 WNQHGLG------GDNVKGSCRDLHPGPVSLLHWSGSGKPWRRLDESKPCPLDALWEPFD 349
W+ GLG D +K + +++H++G+ KPW + ++ PL A + ++
Sbjct: 371 WHVLGLGYNPSISMDEIKNA---------AVVHFNGNMKPWLDIAMTQFKPLWAKYVDYE 523
Query: 350 L 350
L
Sbjct: 524 L 526
>BU494183
Length = 460
Score = 86.7 bits (213), Expect = 6e-18
Identities = 42/126 (33%), Positives = 73/126 (57%)
Frame = +2
Query: 177 VIYLDSDLVLQDDIAKLWNTDLGLNTIGAPQYCHANFTKYFTAAFWSDPVFSTTFEKRKA 236
V++LD D+V+Q D+ LW+TDL N GA + C +F ++ +S+P+ + F+ A
Sbjct: 29 VLFLDDDIVVQKDLTGLWSTDLKGNVNGAVETCGESFHRFDRYLNFSNPLIAKNFDP-PA 205
Query: 237 CYFNTGVMVMDLVKWRKKGYTERIERWMEIQKVERIYELGSLPPFLLVFAGHVAAIEHRW 296
C + G+ V DLV+W+++ TE W ++ ++++LG+LPP L+ F + W
Sbjct: 206 CGWAYGMNVFDLVEWKRQNITEVYHNWQKLNHDRQLWKLGTLPPGLITFWKRTFPLNRYW 385
Query: 297 NQHGLG 302
+ GLG
Sbjct: 386 HVLGLG 403
>AV424294
Length = 395
Score = 81.3 bits (199), Expect = 2e-16
Identities = 47/105 (44%), Positives = 62/105 (58%), Gaps = 2/105 (1%)
Frame = +3
Query: 8 SRFFSAVMTVIIISPSLHSSHYPALAIRSSIIHHIPHHDYRFYFHTSPFFRNADQCEPI- 66
S F V TV I+ S S+ +LA R S I D F +P +RN +C I
Sbjct: 81 SMLFVMVFTVFILLFSSPSNAIRSLANRISDIETEAEVDSFLLFREAPEYRNQQRCTVID 260
Query: 67 -SREIGACHPSLVHVAITLDVEYLRGSIAAVHSILYHASCPENVF 110
+ E C PSLVH+A+ +D EYLRGS+AAVHS+L H+SCP+N+F
Sbjct: 261 TNNENLVCDPSLVHIAMNIDWEYLRGSVAAVHSVLKHSSCPQNLF 395
>BP040145
Length = 530
Score = 80.5 bits (197), Expect = 4e-16
Identities = 48/157 (30%), Positives = 81/157 (51%), Gaps = 10/157 (6%)
Frame = +1
Query: 159 LNYARNYLADLLESCVKRVIYLDSDLVLQDDIAKLWNTDLGLNTIGAPQYCHAN------ 212
LN+ R Y+ +L + + +V++LD D+V+Q D++ LW DL GA + C
Sbjct: 28 LNHLRIYIPELFPN-LDKVVFLDDDVVIQRDLSPLWEIDLEGKVNGAVETCKGEDEWVMS 204
Query: 213 --FTKYFTAAFWSDPVFSTTFEKRKACYFNTGVMVMDLVKWRKKGYTERIERWME--IQK 268
F YF +S P+ + + + C + G+ + DL WR+ E W++ ++
Sbjct: 205 KRFRNYFN---FSHPLVAEHLDPEE-CAWAYGMNIFDLSAWRRTNIRETYHSWLKENLKS 372
Query: 269 VERIYELGSLPPFLLVFAGHVAAIEHRWNQHGLGGDN 305
+++LG+LPP L+ F GHV I+ W+ GLG N
Sbjct: 373 NLTMWKLGTLPPALIAFKGHVHPIDPSWHMLGLGYQN 483
>AW719661
Length = 263
Score = 63.5 bits (153), Expect = 5e-11
Identities = 41/88 (46%), Positives = 54/88 (60%), Gaps = 11/88 (12%)
Frame = +2
Query: 4 IIKFSRFFSAVMTVIIISPSLHSSHYPALAIRSSIIH---HIP------HHDYRFYFHTS 54
+++FS FFSA M +I++SPSL S H PA AIRSS + +P + D RF+F +
Sbjct: 2 LMRFSGFFSAAMLLILLSPSLQSFH-PAEAIRSSHLDGYLRLPVQMPPSNTDDRFFFRQA 178
Query: 55 PFFRNADQCEPISREI--GACHPSLVHV 80
FRNAD+C +R G C PSLVHV
Sbjct: 179NSFRNADECGGSNRMTISGVCDPSLVHV 262
>TC12928
Length = 755
Score = 49.3 bits (116), Expect = 1e-06
Identities = 30/109 (27%), Positives = 54/109 (49%)
Frame = +3
Query: 222 WSDPVFSTTFEKRKACYFNTGVMVMDLVKWRKKGYTERIERWMEIQKVERIYELGSLPPF 281
+SDP + F+ AC + G+ + DL +W++ T +++E+ + GSLP
Sbjct: 54 FSDPFIAKRFDVN-ACTWAFGMNLFDLQQWKRHDLTAVYHKYLEMGAKMPLLNSGSLPLG 230
Query: 282 LLVFAGHVAAIEHRWNQHGLGGDNVKGSCRDLHPGPVSLLHWSGSGKPW 330
L F ++ RW+ GLG D++ + ++ V +H+ G KPW
Sbjct: 231 WLTFYEKTLILDRRWHILGLGYDSIVDT-NEIERAAV--IHYDGIRKPW 368
>TC9237 weakly similar to UP|GLTR_ARATH (Q9FWA4) Probable
glycosyltransferase At3g02350 , partial (4%)
Length = 912
Score = 46.2 bits (108), Expect = 8e-06
Identities = 28/96 (29%), Positives = 48/96 (49%), Gaps = 1/96 (1%)
Frame = +1
Query: 236 ACYFNTGVMVMDLVKWRKKGYTERIERWMEIQKVERIYELGSLPPFLLVFAGHVAAIEHR 295
+C + +G+ ++DLV+WR+ G T+ R ++ ++ G+ LL F + ++
Sbjct: 40 SCAWMSGLNIIDLVRWRELGLTQTYRRLIKEMNIQE--GSGAWRASLLAFENKIYPLDKS 213
Query: 296 WNQHGLGGDNVKGSCRDLHPGPVS-LLHWSGSGKPW 330
W GLG D D H S +LH++G KPW
Sbjct: 214 WVLSGLGHDYT----IDTHAIKTSPVLHYNGKMKPW 309
>AU088971
Length = 218
Score = 43.5 bits (101), Expect = 5e-05
Identities = 19/61 (31%), Positives = 33/61 (53%)
Frame = +3
Query: 242 GVMVMDLVKWRKKGYTERIERWMEIQKVERIYELGSLPPFLLVFAGHVAAIEHRWNQHGL 301
G+ + DL W++K T +W + + +++LG+LPP L+ F G + W+ GL
Sbjct: 6 GMNMFDLKVWKQKDITGIYHKWXNMNEXXVLWKLGTLPPGLMTFYGMTHPLXXSWHVLGL 185
Query: 302 G 302
G
Sbjct: 186 G 188
>CB826753
Length = 494
Score = 27.3 bits (59), Expect = 4.1
Identities = 18/50 (36%), Positives = 24/50 (48%), Gaps = 5/50 (10%)
Frame = -2
Query: 14 VMTVIIISPSLHSS-----HYPALAIRSSIIHHIPHHDYRFYFHTSPFFR 58
++ VI+ S SL S H PA+ RSS IH + HT+P R
Sbjct: 208 IIGVILASKSLPPSLPLVRHPPAMYFRSSSIHFNAFFLIHIHMHTAPKLR 59
>TC14196 homologue to UP|Q9FNV7 (Q9FNV7) Auxin-repressed protein, partial
(95%)
Length = 1040
Score = 27.3 bits (59), Expect = 4.1
Identities = 15/49 (30%), Positives = 23/49 (46%), Gaps = 1/49 (2%)
Frame = -1
Query: 19 IISPSLHSSHYPALAIRSSIIHHIPHHDYRFY-FHTSPFFRNADQCEPI 66
I+ S+ S H+P I +I HH R++ + PFF N C +
Sbjct: 623 ILKRSIGSCHHP--------IQNIAHHHIRYF*LFSFPFFFNLKNCHDV 501
>AV420142
Length = 287
Score = 26.9 bits (58), Expect = 5.3
Identities = 16/51 (31%), Positives = 23/51 (44%)
Frame = +3
Query: 15 MTVIIISPSLHSSHYPALAIRSSIIHHIPHHDYRFYFHTSPFFRNADQCEP 65
+++III H HY +SS IHH +Y H F + Q +P
Sbjct: 144 ISIIIIH---HHYHYQKPQFQSSPIHHHASSSSTWYHHWGSRFPSTPQTKP 287
>TC13397
Length = 478
Score = 26.9 bits (58), Expect = 5.3
Identities = 15/29 (51%), Positives = 20/29 (68%), Gaps = 2/29 (6%)
Frame = -2
Query: 295 RWNQHGLGG-DNVKGSCRDLHP-GPVSLL 321
R+ ++GLGG D G+ DLHP PV+LL
Sbjct: 471 RFVENGLGGGDGGSGTINDLHPLNPVALL 385
>TC13148 similar to UP|Q8LK24 (Q8LK24) SOS2-like protein kinase, partial
(24%)
Length = 545
Score = 26.9 bits (58), Expect = 5.3
Identities = 16/63 (25%), Positives = 25/63 (39%), Gaps = 7/63 (11%)
Frame = -2
Query: 2 SSIIKFSRFFSAVMTVIIISPSLHSSHYPALAIRS-------SIIHHIPHHDYRFYFHTS 54
SS I F ++ + + LH P LA ++HH+ H D+ Y
Sbjct: 544 SSEILLQAAFGDLVEKLAAADELHGDVDPGLAGHDFMEVDDVGVLHHLHHGDFSLYLLHH 365
Query: 55 PFF 57
P+F
Sbjct: 364 PYF 356
>TC17641 homologue to GB|AAP31966.1|30387601|BT006622 At3g19640 {Arabidopsis
thaliana;}, partial (11%)
Length = 521
Score = 26.9 bits (58), Expect = 5.3
Identities = 9/19 (47%), Positives = 15/19 (78%)
Frame = -3
Query: 91 GSIAAVHSILYHASCPENV 109
G+ A+VH I++ A CP+N+
Sbjct: 486 GNAASVHKIMFLAFCPQNI 430
>BP032030
Length = 454
Score = 26.6 bits (57), Expect = 6.9
Identities = 26/89 (29%), Positives = 38/89 (42%), Gaps = 7/89 (7%)
Frame = +3
Query: 25 HSSHYPALAIRSSIIHHIPHH-------DYRFYFHTSPFFRNADQCEPISREIGACHPSL 77
HS+ YPALA R H+ ++ Y+ + P +DQ IS +G CH
Sbjct: 138 HSTLYPALAERQIFFKHVINYFQ*FEIGAYQVHPAPPPQVDKSDQSSAIS--VG-CH--- 299
Query: 78 VHVAITLDVEYLRGSIAAVHSILYHASCP 106
+H + + S+A HS SCP
Sbjct: 300 LHQYFHNHQRH*QSSMAQKHSNGSQQSCP 386
>TC15991 similar to GB|CAB10400.1|2244979|ATFCA5 enoyl-CoA hydratase like
protein {Arabidopsis thaliana;}, partial (57%)
Length = 547
Score = 26.6 bits (57), Expect = 6.9
Identities = 11/24 (45%), Positives = 13/24 (53%)
Frame = +2
Query: 320 LLHWSGSGKPWRRLDESKPCPLDA 343
L WSG+G P L P PLD+
Sbjct: 386 LYKWSGAGNPSSEL*RDSPSPLDS 457
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.326 0.140 0.449
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,926,734
Number of Sequences: 28460
Number of extensions: 132992
Number of successful extensions: 1014
Number of sequences better than 10.0: 47
Number of HSP's better than 10.0 without gapping: 999
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1003
length of query: 360
length of database: 4,897,600
effective HSP length: 91
effective length of query: 269
effective length of database: 2,307,740
effective search space: 620782060
effective search space used: 620782060
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 56 (26.2 bits)
Medicago: description of AC145329.11


