

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC145222.7 - phase: 0
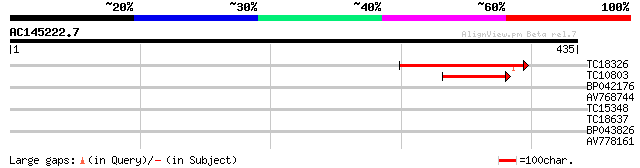
(435 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC18326 weakly similar to UP|O23033 (O23033) YUP8H12.2 protein, ... 170 4e-43
TC10803 similar to UP|O23033 (O23033) YUP8H12.2 protein, partial... 89 1e-18
BP042176 29 1.7
AV768744 28 2.3
TC15348 weakly similar to GB|AAA32913.1|166949|AVOCYP cytochrome... 28 2.3
TC18637 similar to UP|BAD12326 (BAD12326) NADH dehydrogenase sub... 27 5.1
BP043826 27 8.6
AV778161 27 8.6
>TC18326 weakly similar to UP|O23033 (O23033) YUP8H12.2 protein, partial
(19%)
Length = 655
Score = 170 bits (431), Expect = 4e-43
Identities = 83/102 (81%), Positives = 92/102 (89%), Gaps = 3/102 (2%)
Frame = +2
Query: 300 SHIPGFASVLPKLTANLHAMKDKYLKAPHPVSPNTKGKKWDFSFTSIWNTVVWLMLMNFF 359
SHIPGFASVLP + A LHAMKDKYLKAPHPVSPN +GKKWDFSF SIWNTVVWLMLMNFF
Sbjct: 2 SHIPGFASVLPTVIAKLHAMKDKYLKAPHPVSPNIQGKKWDFSFASIWNTVVWLMLMNFF 181
Query: 360 IKIVNSTAQTHLKKQQESEVAALTKK---SDSDTQ*NALIII 398
+KIVN+TAQT+LKKQQE E+AALTKK SDSDTQ*NA +++
Sbjct: 182 VKIVNATAQTYLKKQQERELAALTKKFTTSDSDTQ*NAHVLL 307
>TC10803 similar to UP|O23033 (O23033) YUP8H12.2 protein, partial (11%)
Length = 551
Score = 89.0 bits (219), Expect = 1e-18
Identities = 39/52 (75%), Positives = 47/52 (90%)
Frame = +3
Query: 333 NTKGKKWDFSFTSIWNTVVWLMLMNFFIKIVNSTAQTHLKKQQESEVAALTK 384
N +GKKWDFS S+WNTVVWLMLMNFF+KIVNSTAQ+++KKQQE E+AA T+
Sbjct: 69 NFQGKKWDFSIASVWNTVVWLMLMNFFVKIVNSTAQSYVKKQQEKELAASTE 224
>BP042176
Length = 474
Score = 28.9 bits (63), Expect = 1.7
Identities = 12/27 (44%), Positives = 20/27 (73%)
Frame = +3
Query: 190 AARLSGSRMDAMEELDSEDKGVLNQIK 216
AAR+ G ++EEL +D+GV+NQ++
Sbjct: 132 AARIXGR*KYSLEELHGKDRGVVNQLR 212
>AV768744
Length = 526
Score = 28.5 bits (62), Expect = 2.3
Identities = 12/25 (48%), Positives = 17/25 (68%)
Frame = -2
Query: 217 CWFFSHTQHLNFFTILVLASVPNPL 241
C + T+HL+FFTI VL ++P L
Sbjct: 153 CSIPTRTRHLSFFTIYVLENIPTRL 79
>TC15348 weakly similar to GB|AAA32913.1|166949|AVOCYP cytochrome
P-450LXXIA1 (cyp71A1) {Persea americana;} , partial
(23%)
Length = 614
Score = 28.5 bits (62), Expect = 2.3
Identities = 17/65 (26%), Positives = 29/65 (44%)
Frame = +2
Query: 28 KKTMLYLLAKGGWLMLFSVAVGTLGVVLMTLGCLHEKHLEEFLEYFRFGLWWVALGVASS 87
KK L L+AK +++ V L+ L CL E H++ + Y L+W + +
Sbjct: 419 KKNHLCLVAKNRT*KSL*ISIIQFHVCLLYLSCLQEPHVKLTVTYINATLFWSLIIIMKL 598
Query: 88 IGLGS 92
+ S
Sbjct: 599 FNISS 613
>TC18637 similar to UP|BAD12326 (BAD12326) NADH dehydrogenase subunit 2,
partial (5%)
Length = 506
Score = 27.3 bits (59), Expect = 5.1
Identities = 14/45 (31%), Positives = 24/45 (53%)
Frame = -3
Query: 3 LENLTLTTQPFKTLKYSTLAVIQYIKKTMLYLLAKGGWLMLFSVA 47
L LTL+ PF + + LA+I +I+ + GW+ FS++
Sbjct: 342 LSLLTLSILPFPRVSFFWLAIICFIRGRGSLSMGFTGWVAFFSLS 208
>BP043826
Length = 454
Score = 26.6 bits (57), Expect = 8.6
Identities = 14/36 (38%), Positives = 19/36 (51%)
Frame = +1
Query: 76 GLWWVALGVASSIGLGSGLHTFVLYLGPHIALFTIK 111
GLWWV L +A L LH F L +G + + I+
Sbjct: 343 GLWWVDLWLAGDRLLLYYLHAFPLIIG*GMIYYNIE 450
>AV778161
Length = 548
Score = 26.6 bits (57), Expect = 8.6
Identities = 11/37 (29%), Positives = 21/37 (56%)
Frame = +1
Query: 276 TVFIISVCNNQLLDWIENEFIWVLSHIPGFASVLPKL 312
T++++ +LDW+E + LSH + S+LP +
Sbjct: 430 TLWVMVTIMRTVLDWLEVPYHTELSHCSYYMSLLPSI 540
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.331 0.143 0.461
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,434,382
Number of Sequences: 28460
Number of extensions: 124971
Number of successful extensions: 901
Number of sequences better than 10.0: 16
Number of HSP's better than 10.0 without gapping: 896
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 900
length of query: 435
length of database: 4,897,600
effective HSP length: 93
effective length of query: 342
effective length of database: 2,250,820
effective search space: 769780440
effective search space used: 769780440
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.9 bits)
S2: 57 (26.6 bits)
Medicago: description of AC145222.7


