

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC145222.4 - phase: 0
(521 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
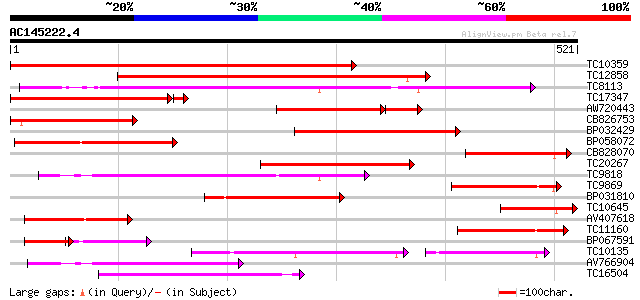
Score E
Sequences producing significant alignments: (bits) Value
TC10359 similar to UP|Q94AZ2 (Q94AZ2) AT5g26340/F9D12_17, partia... 553 e-158
TC12858 weakly similar to UP|Q9ZS76 (Q9ZS76) Hexose transporter,... 277 3e-75
TC8113 weakly similar to UP|O23213 (O23213) Sugar transporter li... 192 9e-50
TC17347 similar to UP|Q9FMX3 (Q9FMX3) Monosaccharide transporter... 179 5e-47
AW720443 146 1e-45
CB826753 179 1e-45
BP032429 161 3e-40
BP058072 158 2e-39
CB828070 151 3e-37
TC20267 similar to UP|Q7XA52 (Q7XA52) Monosaccharide transporter... 144 3e-35
TC9818 similar to UP|Q7XA50 (Q7XA50) Sorbitol-like transporter, ... 134 4e-32
TC9869 similar to UP|O65413 (O65413) Glucose transporter (STP12 ... 134 5e-32
BP031810 115 2e-26
TC10645 similar to UP|Q41139 (Q41139) Hexose carrier (Fragment),... 114 3e-26
AV407618 113 9e-26
TC11160 similar to UP|Q7XA51 (Q7XA51) Monosaccharide transporter... 111 3e-25
BP067591 72 2e-24
TC10135 108 2e-24
AV766904 96 1e-20
TC16504 weakly similar to UP|Q84KI7 (Q84KI7) Sorbitol transporte... 87 5e-18
>TC10359 similar to UP|Q94AZ2 (Q94AZ2) AT5g26340/F9D12_17, partial (90%)
Length = 1099
Score = 553 bits (1426), Expect = e-158
Identities = 278/318 (87%), Positives = 295/318 (92%)
Frame = +2
Query: 1 MAGGGFATSGGGEFEAKITPIVIISCIMAATGGLMFGYDVGVSGGVTSMHPFLKKFFPAV 60
MAGGGFATSGGG+FEAKITPIVIISCI+AATGGLMFGYDVGVSGGVTSM FL KFFP V
Sbjct: 146 MAGGGFATSGGGDFEAKITPIVIISCILAATGGLMFGYDVGVSGGVTSMPHFLHKFFPDV 325
Query: 61 YRKTVLEAGLDSNYCKYDNQGLQLFTSSLYLAALTSTFFASYTTRTMGRRLTMLIAGFFF 120
Y+KTVLE LDSNYCKYDNQGLQLFTSSLYLA L STFFASYTTRT+GRR+TMLIAG FF
Sbjct: 326 YKKTVLEKELDSNYCKYDNQGLQLFTSSLYLAGLVSTFFASYTTRTLGRRMTMLIAGVFF 505
Query: 121 IAGVAFNAAAQNLAMLIVGRILLGCGVGFANQAVPVFLSEIAPSRIRGALNILFQLNVTI 180
IAGV FN AQNLAMLIVGRILLGCGVGFANQAVP+FLSEIAPSRIRGALNILFQLN+TI
Sbjct: 506 IAGVVFNVTAQNLAMLIVGRILLGCGVGFANQAVPLFLSEIAPSRIRGALNILFQLNITI 685
Query: 181 GILFANLVNYGTNKISGGWGWRLSLGLAGIPALLLTVGAIVVVDTPNSLIERGRLEEGKA 240
GILFANL+NYGTNKI GGWGWRLSLGLAGIPA+LLTVGA++VVDTPNSLIERGR EEGKA
Sbjct: 686 GILFANLINYGTNKIKGGWGWRLSLGLAGIPAVLLTVGALLVVDTPNSLIERGRTEEGKA 865
Query: 241 VLKKIRGTDNIEPEFLELCEASRVAKEVKQAFRNLLKGEKGGQVIMAIGLEIFQQFTGIN 300
VLKKIRGTDN+EPEFLEL EASRVAKEVK FRNLLK Q+I++I L+IFQQFTGIN
Sbjct: 866 VLKKIRGTDNVEPEFLELVEASRVAKEVKDPFRNLLKRRNRPQIIISIALQIFQQFTGIN 1045
Query: 301 AIMFYAPVLFNTVGFKND 318
AIMFYAPVLFNT+GFKND
Sbjct: 1046AIMFYAPVLFNTLGFKND 1099
>TC12858 weakly similar to UP|Q9ZS76 (Q9ZS76) Hexose transporter, partial
(51%)
Length = 882
Score = 277 bits (709), Expect = 3e-75
Identities = 143/289 (49%), Positives = 194/289 (66%), Gaps = 2/289 (0%)
Frame = +3
Query: 100 ASYTTRTMGRRLTMLIAGFFFIAGVAFNAAAQNLAMLIVGRILLGCGVGFANQAVPVFLS 159
AS TR MG R TM++ G FF++G FN A + MLIVGR+LLG G+G ANQ+VP++LS
Sbjct: 12 ASTITRIMGMRATMIVGGLFFVSGALFNGLADGIWMLIVGRLLLGFGIGCANQSVPIYLS 191
Query: 160 EIAPSRIRGALNILFQLNVTIGILFANLVNYGTNKISGGWGWRLSLGLAGIPALLLTVGA 219
E+AP + RG LN+ FQL++TIGI ANL NY KI G GWRLSLGL +PA++ VG+
Sbjct: 192 EMAPYKYRGGLNMCFQLSITIGIFVANLFNYYFAKILNGQGWRLSLGLGAVPAVIFVVGS 371
Query: 220 IVVVDTPNSLIERGRLEEGKAVLKKIRGTDNIEPEFLELCEASRVAKEVKQAFRNLLKGE 279
+ + D+P+SL+ RGR E + L KIRGT +IE E ++ AS + VK ++ LL+ +
Sbjct: 372 LCLPDSPSSLVARGRHEAARQELVKIRGTTDIEAELKDIITASEALENVKHPWKTLLERK 551
Query: 280 KGGQVIMAIGLEIFQQFTGINAIMFYAPVLFNTVGFKNDASLYSAVITGAVNVLSTIVSI 339
Q++ A+ + FQQFTG+N I FYAP+LF T+GF ASL SAVI G+ +ST++SI
Sbjct: 552 YRPQLVFAVCIPFFQQFTGLNVITFYAPILFRTIGFGPTASLMSAVIIGSFKPVSTLISI 731
Query: 340 YFVDKLGRRMLLLEAGVQMFLSQIV--IAIILGIKVTDHSDDLSKGYAI 386
+ VDK GRR L LE G QM + QI+ IAI + + + L K YA+
Sbjct: 732 FVVDKFGRRTLFLEGGAQMLICQIIMTIAIAVTFGTSGNPGQLPKWYAV 878
>TC8113 weakly similar to UP|O23213 (O23213) Sugar transporter like
protein, partial (93%)
Length = 1865
Score = 192 bits (489), Expect = 9e-50
Identities = 139/497 (27%), Positives = 234/497 (46%), Gaps = 23/497 (4%)
Frame = +2
Query: 10 GGGEFEAKITPIVIISCIMAATGGLMFGYDVGVSGGVTSMHPFLKKFFPAVYRKTVLEAG 69
G + + I + I+A+ ++ GYD GV G F+K+ + G
Sbjct: 80 GNEDKDTTINKYALACVIVASMVSIISGYDTGVMSGALL---FIKE-----------DIG 217
Query: 70 LDSNYCKYDNQGLQLFTSSLYLAALTSTFFASYTTRTMGRRLTMLIAGFFFIAGVAFNAA 129
+ D Q ++ L + AL A T+ +GRR T+ +A F+ G F
Sbjct: 218 IS------DTQQ-EVLAGILNICALVGCLAAGKTSDYIGRRYTIFLASILFLVGAVFMGY 376
Query: 130 AQNLAMLIVGRILLGCGVGFANQAVPVFLSEIAPSRIRGALNILFQLNVTIGILFANLVN 189
N A+L+ GR + G GVGFA PV+ +E++ + RG L L ++ + +GI + N
Sbjct: 377 GPNFAILMFGRCVCGLGVGFALTTAPVYSAELSSASTRGFLTSLPEVCIGLGIFIGYISN 556
Query: 190 YGTNKISGGWGWRLSLGLAGIPALLLTVGAIVVVDTPNSLIERGRLEEGKAVLKKIRG-T 248
Y K++ GWRL LGLA IP+L L +G + + ++P L+ +GRL K VL ++ T
Sbjct: 557 YFLGKLALTLGWRLMLGLAAIPSLGLALGILTMPESPRWLVMQGRLGCAKKVLLEVSNTT 736
Query: 249 DNIEPEFLELCEASRVAKEVKQAFRNLLKGEKGGQ-----------------VIMAIGLE 291
+ E F ++ ++ ++ F + G+ +I A+G+
Sbjct: 737 EEAELRFRDIIVSAGFDEKCNDEFVKQPQKSHHGEGVWKELFLRPTPPVRRMLIAAVGIH 916
Query: 292 IFQQFTGINAIMFYAPVLFNTVGF-KNDASLYSAVITGAVNVLSTIVSIYFVDKLGRRML 350
F+ TGI A+M Y P +F G D L + + TG + +S + +D++GRR L
Sbjct: 917 FFEHATGIEAVMLYGPRIFKKAGVTTKDRLLLATIGTGLTKITFLTISTFLLDRVGRRRL 1096
Query: 351 LLEAGVQMFLSQIVIAI-ILGIKVT--DHSDDLSKGYAIFVVILVCTFVSAFAWSWGPLG 407
L Q+ ++ ++ + ILG +T ++S + ++ T+V+ F P+
Sbjct: 1097L-----QISVAGMIFGLTILGFSLTMVEYSSEKLVWALSLSIVATYTYVAFFNVGLAPVT 1261
Query: 408 WLIPSETFPLETRSAGQSVTVCVNMLFTFVIAQAFLSMLCHFKF-GIFLFFSGWVLIMSI 466
W+ SE FPL R+ G S+ V VN I+ +F+S+ G F F+G ++ +
Sbjct: 1262WVYSSEIFPLRLRAQGNSIGVAVNRGMNAAISMSFISIYKAITIGGAFFLFAGMSVVAWV 1441
Query: 467 FVLFLVPETKNIPIEEM 483
F F +PETK +EEM
Sbjct: 1442FFYFCLPETKGKALEEM 1492
>TC17347 similar to UP|Q9FMX3 (Q9FMX3) Monosaccharide transporter (STP11
protein), partial (30%)
Length = 663
Score = 179 bits (453), Expect(2) = 5e-47
Identities = 85/150 (56%), Positives = 112/150 (74%), Gaps = 1/150 (0%)
Frame = +3
Query: 1 MAGGGFATSGGG-EFEAKITPIVIISCIMAATGGLMFGYDVGVSGGVTSMHPFLKKFFPA 59
MAGG + SG EF+ K+T V+++C +AA GGL+FGYD+G++GGVTSM PFL KFFP
Sbjct: 168 MAGGAYVDSGKAKEFDGKVTAFVLVTCFVAAMGGLLFGYDLGITGGVTSMEPFLVKFFPG 347
Query: 60 VYRKTVLEAGLDSNYCKYDNQGLQLFTSSLYLAALTSTFFASYTTRTMGRRLTMLIAGFF 119
VY++ E+G +S YCK+DN+ L LFTSSLYLAAL ++FFAS TTR +GR+ +M G F
Sbjct: 348 VYKQMKDESGHESEYCKFDNELLTLFTSSLYLAALIASFFASTTTRMLGRKTSMFAGGLF 527
Query: 120 FIAGVAFNAAAQNLAMLIVGRILLGCGVGF 149
F+ G N A N+ MLI+GR+LLG GVG+
Sbjct: 528 FLVGALLNGFAVNIEMLIIGRLLLGFGVGY 617
Score = 26.2 bits (56), Expect(2) = 5e-47
Identities = 10/14 (71%), Positives = 14/14 (99%)
Frame = +2
Query: 151 NQAVPVFLSEIAPS 164
NQ+VPV+LSE+AP+
Sbjct: 620 NQSVPVYLSEMAPA 661
>AW720443
Length = 406
Score = 146 bits (368), Expect(2) = 1e-45
Identities = 74/100 (74%), Positives = 86/100 (86%)
Frame = +2
Query: 246 RGTDNIEPEFLELCEASRVAKEVKQAFRNLLKGEKGGQVIMAIGLEIFQQFTGINAIMFY 305
RGTDNIE EF +L EA+RVAK+VK FRN+LK + Q+++AI L+IFQQFTGINAIMFY
Sbjct: 2 RGTDNIELEFSDLVEATRVAKQVKHPFRNILKRKNRPQLVIAIALQIFQQFTGINAIMFY 181
Query: 306 APVLFNTVGFKNDASLYSAVITGAVNVLSTIVSIYFVDKL 345
APVLFNT GFKNDA+LYSAVI GAVN +STIVSIY VD+L
Sbjct: 182 APVLFNTFGFKNDAALYSAVIVGAVNCVSTIVSIYSVDRL 301
Score = 54.7 bits (130), Expect(2) = 1e-45
Identities = 26/34 (76%), Positives = 30/34 (87%)
Frame = +3
Query: 346 GRRMLLLEAGVQMFLSQIVIAIILGIKVTDHSDD 379
GRRMLLLEAG QM LS +VIA++LGIKV DHS+D
Sbjct: 303 GRRMLLLEAGFQMLLSLMVIAVVLGIKVKDHSED 404
>CB826753
Length = 494
Score = 179 bits (454), Expect = 1e-45
Identities = 92/119 (77%), Positives = 100/119 (83%), Gaps = 2/119 (1%)
Frame = +3
Query: 1 MAGGGFATS--GGGEFEAKITPIVIISCIMAATGGLMFGYDVGVSGGVTSMHPFLKKFFP 58
MAGG + GG +FEAKITPI+IISCIMAATGGLMFGYDVGVSGGVTSM FLKKFFP
Sbjct: 138 MAGGCLTSGSDGGRDFEAKITPIIIISCIMAATGGLMFGYDVGVSGGVTSMPHFLKKFFP 317
Query: 59 AVYRKTVLEAGLDSNYCKYDNQGLQLFTSSLYLAALTSTFFASYTTRTMGRRLTMLIAG 117
V+RKT E +SNYCKYDNQ LQLFTSSLYLA LT TFFASYTTR +GRRLTML++G
Sbjct: 318 EVFRKTKEEEEFNSNYCKYDNQMLQLFTSSLYLAGLTVTFFASYTTRRLGRRLTMLVSG 494
>BP032429
Length = 468
Score = 161 bits (407), Expect = 3e-40
Identities = 78/154 (50%), Positives = 107/154 (68%), Gaps = 1/154 (0%)
Frame = -2
Query: 262 SRVAKEVKQAFRNLLKGEKGGQVIMAIGLEIFQQFTGINAIMFYAPVLFNTVGFKNDASL 321
S +K+V+ ++N+ K Q++ + FQQ TGIN IMFYAPVLF T+GF +DA+L
Sbjct: 467 SEASKKVEHPWKNITKPVYKPQMVFCSLIPFFQQLTGINVIMFYAPVLFKTLGFGDDAAL 288
Query: 322 YSAVITGAVNVLSTIVSIYFVDKLGRRMLLLEAGVQMFLSQIVIAIILGIKV-TDHSDDL 380
SAVI+G VNVL+T VSI+ VDK GRR+L LE GVQM +SQI + ++ +K
Sbjct: 287 MSAVISGGVNVLATFVSIFSVDKFGRRLLFLEGGVQMLISQIAVGTMIALKFGVSGEGSF 108
Query: 381 SKGYAIFVVILVCTFVSAFAWSWGPLGWLIPSET 414
+KG A ++ +C +V+AFAWSWGPLGWL+PSE+
Sbjct: 107 TKGEANLLLFFICAYVAAFAWSWGPLGWLVPSES 6
>BP058072
Length = 520
Score = 158 bits (399), Expect = 2e-39
Identities = 80/150 (53%), Positives = 102/150 (67%)
Frame = -1
Query: 5 GFATSGGGEFEAKITPIVIISCIMAATGGLMFGYDVGVSGGVTSMHPFLKKFFPAVYRKT 64
G GG E+ +T V ++CI+AA GGL+FGYD+G+SGGVTSM PFL+KFF +VY K
Sbjct: 520 GIGNGGGKEYTGYLTRFVTVTCIVAAMGGLIFGYDIGISGGVTSMDPFLRKFFHSVYLKK 341
Query: 65 VLEAGLDSNYCKYDNQGLQLFTSSLYLAALTSTFFASYTTRTMGRRLTMLIAGFFFIAGV 124
E + YC+YD+Q L +FTSSLYLAAL S+ AS TR +L+M + G F+
Sbjct: 340 Q-EKSSPNKYCQYDSQVLTIFTSSLYLAALLSSLVASTITRRFSPKLSMFVGGMHFVVVA 164
Query: 125 AFNAAAQNLAMLIVGRILLGCGVGFANQAV 154
N AQ + MLIVGRILLG G+GFANQA+
Sbjct: 163 LLNGLAQRVWMLIVGRILLGIGIGFANQAL 74
>CB828070
Length = 530
Score = 151 bits (381), Expect = 3e-37
Identities = 69/101 (68%), Positives = 83/101 (81%), Gaps = 4/101 (3%)
Frame = +1
Query: 420 RSAGQSVTVCVNMLFTFVIAQAFLSMLCHFKFGIFLFFSGWVLIMSIFVLFLVPETKNIP 479
RSAGQS+TVCVN+ FTF+IAQAFLSM+C KFGIF+FFSGW LIMS+FV FL+PETKN+P
Sbjct: 1 RSAGQSLTVCVNLAFTFLIAQAFLSMMCSLKFGIFIFFSGWALIMSVFVFFLLPETKNVP 180
Query: 480 IEEMTERVWKQHWFWKRFME----DDNEKVSNADYPKIKNN 516
IEEM ERVWKQHW WKRF+E +D++KV++ P N
Sbjct: 181 IEEMVERVWKQHWLWKRFVENDYVEDDQKVADGKGPTRNEN 303
>TC20267 similar to UP|Q7XA52 (Q7XA52) Monosaccharide transporter, partial
(31%)
Length = 525
Score = 144 bits (364), Expect = 3e-35
Identities = 75/142 (52%), Positives = 100/142 (69%)
Frame = +2
Query: 231 ERGRLEEGKAVLKKIRGTDNIEPEFLELCEASRVAKEVKQAFRNLLKGEKGGQVIMAIGL 290
+R R E ++RG D+++ EF +L EAS + +V+ +RNLL+ + + MAI +
Sbjct: 56 DRTRXPERSGSS*RVRGVDDVDEEFSDLVEASEASMQVEHPWRNLLQRKYRPHLTMAILI 235
Query: 291 EIFQQFTGINAIMFYAPVLFNTVGFKNDASLYSAVITGAVNVLSTIVSIYFVDKLGRRML 350
FQQFTGIN IMFYAPVLF+++GFK+DASL SAVITG VNV++T VSIY VDK GRR L
Sbjct: 236 PFFQQFTGINVIMFYAPVLFSSIGFKDDASLMSAVITGVVNVVATCVSIYGVDKWGRRKL 415
Query: 351 LLEAGVQMFLSQIVIAIILGIK 372
L+ GVQM + Q V+A +G K
Sbjct: 416 FLQGGVQMMICQAVVAAAIGAK 481
>TC9818 similar to UP|Q7XA50 (Q7XA50) Sorbitol-like transporter, partial
(62%)
Length = 1162
Score = 134 bits (337), Expect = 4e-32
Identities = 90/324 (27%), Positives = 155/324 (47%), Gaps = 20/324 (6%)
Frame = +2
Query: 27 IMAATGGLMFGYDVGVSGGVTSMHPFLKKFFPAVYRKTVLEAGLDSNYCKYDNQGLQLFT 86
++A+ ++ GYD+GV G A+Y K L K + +++
Sbjct: 251 MLASMTSILLGYDIGVMSGA------------AIYIKRDL---------KVSDVKIEILL 367
Query: 87 SSLYLAALTSTFFASYTTRTMGRRLTMLIAGFFFIAGVAFNAAAQNLAMLIVGRILLGCG 146
+ L +L + A T+ +GRR T++ AG F G + N L+ GR + G G
Sbjct: 368 GIINLYSLIGSGLAGRTSDWIGRRYTIVFAGAIFFVGALLMGFSPNYWFLMFGRFIAGIG 547
Query: 147 VGFANQAVPVFLSEIAPSRIRGALNILFQLNVTIGILFANLVNYGTNKISGGWGWRLSLG 206
+G+A PV+ +E++P+ RG L ++ + GIL + N+ +K+S GWR+ LG
Sbjct: 548 IGYALMIAPVYTAEVSPASSRGFLTSFPEVFINGGILLGYISNFAFSKLSLKVGWRMMLG 727
Query: 207 LAGIPALLLTVGAIVVVDTPNSLIERGRLEEGKAVLKKIRGTDNIEPEFLELCEASRVAK 266
+ +P+++L VG + + ++P L+ RGRL + VL K +D+ E L L + R A
Sbjct: 728 VGALPSVILGVGVLAMPESPRWLVMRGRLGDAIKVLNKT--SDSPEEAQLRLADIKRAAG 901
Query: 267 EVKQAFRNLLKGEKGGQ-------------------VIMAIGLEIFQQFTGINAIMFYAP 307
+ ++++ K VI A+G+ FQQ +GI+A++ Y+P
Sbjct: 902 IPESCTDDVVEVSKRSTGEGVWKELFLYPTPAIRHIVIAALGIHFFQQASGIDAVVLYSP 1081
Query: 308 VLFNTVGFKNDA-SLYSAVITGAV 330
+F G K+D L + V G V
Sbjct: 1082TIFEKAGIKSDTDKLLATVAVGFV 1153
>TC9869 similar to UP|O65413 (O65413) Glucose transporter (STP12 protein),
partial (18%)
Length = 544
Score = 134 bits (336), Expect = 5e-32
Identities = 62/103 (60%), Positives = 77/103 (74%), Gaps = 2/103 (1%)
Frame = +1
Query: 407 GWLIPSETFPLETRSAGQSVTVCVNMLFTFVIAQAFLSMLCHFKFGIFLFFSGWVLIMSI 466
GWL+PSE LE RSAGQ++ V VNMLFTF IAQ FLSMLCH KFG+F FF+G+V+IM+I
Sbjct: 13 GWLVPSEICSLEVRSAGQALNVAVNMLFTFAIAQIFLSMLCHLKFGLFFFFAGFVVIMTI 192
Query: 467 FVLFLVPETKNIPIEEMTERVWKQHWFWKRFM--EDDNEKVSN 507
FV +PETK +PIEEM VW+ HWFW + + D+N+ N
Sbjct: 193 FVALFLPETKGVPIEEMNS-VWRSHWFWSKIVPAADNNDNRKN 318
>BP031810
Length = 393
Score = 115 bits (287), Expect = 2e-26
Identities = 59/129 (45%), Positives = 88/129 (67%), Gaps = 1/129 (0%)
Frame = +3
Query: 180 IGILFANLVNYGTNKISGGWGWRLSLGLAGIPALLLTVGAIVVVDTPNSLIERGRLEEGK 239
+G ANLVNY T ++ WGWRLSLGLA +PA ++ +G ++ +TPNSL+E+GRLEE +
Sbjct: 9 LGNFDANLVNYATEQLHP-WGWRLSLGLATVPATVMFIGGLLCPETPNSLVEQGRLEEAR 185
Query: 240 AVLKKIRGTDNIEPEFLELCEASRVAKEVKQAFRN-LLKGEKGGQVIMAIGLEIFQQFTG 298
VL+++RGT N++ E+ ++ EAS A+++K F+N LLK + VI A+ + FQQ TG
Sbjct: 186 QVLERVRGTPNVDAEYEDIVEASVEAQKIKNPFQNLLLKKNRPQFVIGALAIPAFQQLTG 365
Query: 299 INAIMFYAP 307
N+ P
Sbjct: 366 NNSHPLLCP 392
>TC10645 similar to UP|Q41139 (Q41139) Hexose carrier (Fragment), partial
(47%)
Length = 524
Score = 114 bits (286), Expect = 3e-26
Identities = 55/73 (75%), Positives = 61/73 (83%), Gaps = 3/73 (4%)
Frame = -2
Query: 452 GIFLFFSGWVLIMSIFVLFLVPETKNIPIEEMTERVWKQHWFWKRFMEDD---NEKVSNA 508
GIFLFFSGWVLIMS+FV FLVPETKN+PIEEMTERVWKQHWFWKR+MEDD NEKV+N
Sbjct: 523 GIFLFFSGWVLIMSLFVYFLVPETKNVPIEEMTERVWKQHWFWKRYMEDDYVANEKVANV 344
Query: 509 DYPKIKNNPNSQL 521
+ +P SQL
Sbjct: 343 NGTTKGFDPVSQL 305
>AV407618
Length = 334
Score = 113 bits (282), Expect = 9e-26
Identities = 55/100 (55%), Positives = 72/100 (72%)
Frame = +2
Query: 14 FEAKITPIVIISCIMAATGGLMFGYDVGVSGGVTSMHPFLKKFFPAVYRKTVLEAGLDSN 73
+E + TP +C + A GG +FGYD+GVSGGVTSM FLK+FFP VYR+ +++
Sbjct: 38 YEHRFTPYFAFTCFVGALGGSLFGYDLGVSGGVTSMDDFLKEFFPKVYRRKHTHL-KETD 214
Query: 74 YCKYDNQGLQLFTSSLYLAALTSTFFASYTTRTMGRRLTM 113
YCKYD+Q L LFTSSLY++AL TFFASY TR GR+ ++
Sbjct: 215 YCKYDDQILTLFTSSLYISALFMTFFASYLTRNKGRKASI 334
>TC11160 similar to UP|Q7XA51 (Q7XA51) Monosaccharide transporter, partial
(16%)
Length = 538
Score = 111 bits (278), Expect = 3e-25
Identities = 51/102 (50%), Positives = 72/102 (70%)
Frame = +1
Query: 412 SETFPLETRSAGQSVTVCVNMLFTFVIAQAFLSMLCHFKFGIFLFFSGWVLIMSIFVLFL 471
SE FPLE RSA QS+ VCVNM FT ++AQ FL LC+ K+GIFL F G +++MS F+ FL
Sbjct: 1 SELFPLEIRSAAQSIVVCVNMTFTALVAQLFLMSLCNLKYGIFLLFGGLIVVMSCFIFFL 180
Query: 472 VPETKNIPIEEMTERVWKQHWFWKRFMEDDNEKVSNADYPKI 513
+PETK +PIEE+ +++ HWFWK+ + + + ++ P I
Sbjct: 181 LPETKQVPIEEI-YLLFENHWFWKKIVRANQDFEASNGKPAI 303
>BP067591
Length = 358
Score = 71.6 bits (174), Expect(2) = 2e-24
Identities = 30/45 (66%), Positives = 42/45 (92%)
Frame = +2
Query: 14 FEAKITPIVIISCIMAATGGLMFGYDVGVSGGVTSMHPFLKKFFP 58
F++KIT ++I+CI+AA+GGL+FGYDVG++GGVT+M PFL+KFFP
Sbjct: 11 FDSKITLAILITCIVAASGGLIFGYDVGIAGGVTTMLPFLEKFFP 145
Score = 58.2 bits (139), Expect(2) = 2e-24
Identities = 35/80 (43%), Positives = 47/80 (58%), Gaps = 1/80 (1%)
Frame = +3
Query: 52 FLKKFFPAVYRKTVLEAGLDSN-YCKYDNQGLQLFTSSLYLAALTSTFFASYTTRTMGRR 110
FLK ++ RK AG D N YC YD+Q L LFTSSLYLA L ++ AS +GR+
Sbjct: 126 FLKNSSRSILRKA---AGSDVNMYCVYDSQVLTLFTSSLYLAGLVTSLLASRVRAWIGRK 296
Query: 111 LTMLIAGFFFIAGVAFNAAA 130
T+++ G F+ G A A+
Sbjct: 297 NTIILGGATFLGGGAVKGAS 356
>TC10135
Length = 1501
Score = 108 bits (270), Expect = 2e-24
Identities = 71/207 (34%), Positives = 112/207 (53%), Gaps = 8/207 (3%)
Frame = +2
Query: 168 GALNILFQLNVTIGILFANLVNYGTNKISGGWGWRLSLGLAGIPALLLTVGAIVVVDTPN 227
GAL L +T G + L+N G W W L G+A PA++ V + + ++P
Sbjct: 2 GALVSLNSFLITGGQFLSYLINLAFTNAPGTWRWML--GVAAAPAIIQIVLMLSLPESPR 175
Query: 228 SLIERGRLEEGKAVLKKIRGTDNIEPEFLELCEA----SRVAKEVKQAFRNLLKGEKGGQ 283
L +GR EE K++LKKI ++++ E L E+ +K K + LLK +
Sbjct: 176 WLYRKGREEESKSILKKIYAPEDVDAEIEALKESVESEIEESKTSKISMMTLLKTTTVRR 355
Query: 284 VIMA-IGLEIFQQFTGINAIMFYAPVLFNTVGF-KNDASLYSAVITGAVNVLSTIVSIYF 341
+ A +GL+ FQQF GIN +M+Y+P + GF N +L ++IT +N +I+SIYF
Sbjct: 356 GLYAGMGLQFFQQFVGINTVMYYSPTIVQLAGFASNRTALLLSLITSGLNAFGSILSIYF 535
Query: 342 VDKLGRRMLLLEA--GVQMFLSQIVIA 366
+DK GR+ L L + GV + L+ + +A
Sbjct: 536 IDKTGRKKLALISLCGVVLSLALLTVA 616
Score = 50.4 bits (119), Expect = 7e-07
Identities = 32/118 (27%), Positives = 57/118 (48%), Gaps = 4/118 (3%)
Frame = +2
Query: 383 GYAIFVVILVCTFVSAFAWSWGPLGWLIPSETFPLETRSAGQSVTVCVNMLFTFVIAQAF 442
G+A V + + ++ F+ G + W+I SE +P R + + +++Q+F
Sbjct: 884 GWAALVGLAL--YIITFSPGMGTVPWVINSEIYPFRYRGVCGGIASTTVWISNLIVSQSF 1057
Query: 443 LSMLCHFKFG-IFLFFSGWVLIMSIFVLFLVPETKNIPIEE---MTERVWKQHWFWKR 496
LS+ F+ F ++ FV+ VPETK +P+EE M E+ Q FW++
Sbjct: 1058LSLTQTIGTAWTFMMFGILAVVAIFFVIVFVPETKGVPMEEVEKMLEQRSLQFKFWQK 1231
>AV766904
Length = 611
Score = 95.9 bits (237), Expect = 1e-20
Identities = 58/199 (29%), Positives = 96/199 (48%)
Frame = +3
Query: 17 KITPIVIISCIMAATGGLMFGYDVGVSGGVTSMHPFLKKFFPAVYRKTVLEAGLDSNYCK 76
K + I+A+ ++ GYD+GV G +Y K L K
Sbjct: 75 KRNKFALACAILASMTSILLGYDIGVMSGAV------------IYIKRDL---------K 191
Query: 77 YDNQGLQLFTSSLYLAALTSTFFASYTTRTMGRRLTMLIAGFFFIAGVAFNAAAQNLAML 136
+ +++ + + + + ++ A T+ +GRR T+++AG F G + N A L
Sbjct: 192 ISDVQIEILSGIMNIYSPIGSYIAGRTSDWIGRRYTIVLAGLIFFVGAILMGFSPNYAFL 371
Query: 137 IVGRILLGCGVGFANQAVPVFLSEIAPSRIRGALNILFQLNVTIGILFANLVNYGTNKIS 196
+ GR + G G+GFA PV+ SEI+P+ RG L L ++ + GIL + NY +K+
Sbjct: 372 MFGRFVAGVGIGFAFLIAPVYTSEISPTLSRGFLTSLPEVFLNAGILIGYISNYTFSKLP 551
Query: 197 GGWGWRLSLGLAGIPALLL 215
GWRL LG+ IP++ L
Sbjct: 552 LRLGWRLMLGIGAIPSIFL 608
>TC16504 weakly similar to UP|Q84KI7 (Q84KI7) Sorbitol transporter, partial
(37%)
Length = 903
Score = 87.4 bits (215), Expect = 5e-18
Identities = 54/190 (28%), Positives = 92/190 (48%)
Frame = +2
Query: 82 LQLFTSSLYLAALTSTFFASYTTRTMGRRLTMLIAGFFFIAGVAFNAAAQNLAMLIVGRI 141
+Q L+++AL + A +GRR T+++ F+ G + +L++G
Sbjct: 203 VQFSVGLLHMSALPACMTAGRIADYIGRRYTIILTAIIFLVGSVLMGYGPSYPILMIGLS 382
Query: 142 LLGCGVGFANQAVPVFLSEIAPSRIRGALNILFQLNVTIGILFANLVNYGTNKISGGWGW 201
+ G G+GF+ PV+ +EI+P RG L +++ GI + NY K+S GW
Sbjct: 383 ISGIGLGFSLIVAPVYCAEISPPSHRGFLTSFLEVSTNAGIALGYVSNYFFGKLSIRLGW 562
Query: 202 RLSLGLAGIPALLLTVGAIVVVDTPNSLIERGRLEEGKAVLKKIRGTDNIEPEFLELCEA 261
RL L + +P+L L + V++P L+ +GR+ E + VL + T EA
Sbjct: 563 RLMLVVHAVPSLALVALMLNWVESPRWLVMQGRVGEARKVLLLVSNTKE---------EA 715
Query: 262 SRVAKEVKQA 271
+ KE+K A
Sbjct: 716 EQRLKEIKVA 745
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.326 0.142 0.425
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,428,781
Number of Sequences: 28460
Number of extensions: 113822
Number of successful extensions: 1080
Number of sequences better than 10.0: 83
Number of HSP's better than 10.0 without gapping: 1043
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1053
length of query: 521
length of database: 4,897,600
effective HSP length: 94
effective length of query: 427
effective length of database: 2,222,360
effective search space: 948947720
effective search space used: 948947720
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 57 (26.6 bits)
Medicago: description of AC145222.4


