

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC145222.13 + phase: 0
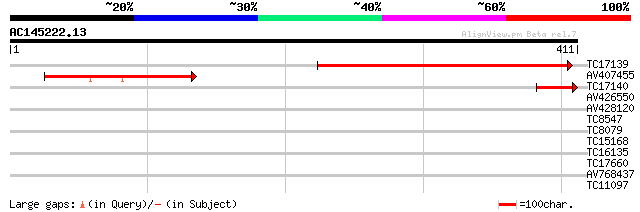
(411 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC17139 332 9e-92
AV407455 113 7e-26
TC17140 similar to GB|AAB98753.1|1591472|U67521 C4-dicarboxylate... 54 6e-08
AV426550 32 0.25
AV428120 29 1.3
TC8547 homologue to UP|MMK1_MEDSA (Q07176) Mitogen-activated pro... 29 1.6
TC8079 homologue to UP|Q9M6S1 (Q9M6S1) MAP kinase 3, complete 28 3.7
TC15168 similar to UP|O04084 (O04084) Serine carboxypeptidase is... 27 4.8
TC16135 UP|PSAB_LOTJA (P58385) Photosystem I P700 chlorophyll A ... 27 4.8
TC17660 homologue to UP|Q9M6R8 (Q9M6R8) MAP kinase PsMAPK2, part... 27 6.2
AV768437 27 8.1
TC11097 similar to UP|P93520 (P93520) Calcium/calmodulin-depende... 27 8.1
>TC17139
Length = 556
Score = 332 bits (850), Expect = 9e-92
Identities = 162/185 (87%), Positives = 175/185 (94%)
Frame = +1
Query: 224 LQGALRGLAYMHDHDRLHQSLGPFSVSLNTISESEAPYLIPRLRDLAFSVSVRYSELEDS 283
L GA+RGLAYMHDH+RLHQSLGPFSV+LNTISE EA YLIPRLRDLAFSV +RYSELE+S
Sbjct: 1 LHGAMRGLAYMHDHERLHQSLGPFSVALNTISEREARYLIPRLRDLAFSVDIRYSELENS 180
Query: 284 GPLTEGLWARASAASAFTYLEKRAFGIADDIYEAGLLFAYLAFVPFCEAGVMDGLSLQRL 343
G L +GLWARAS A AFTYLEKRAFGIADDIYEAGLLFAYLAFVPFCEAGV DGLSLQRL
Sbjct: 181 GSLADGLWARASRAGAFTYLEKRAFGIADDIYEAGLLFAYLAFVPFCEAGVTDGLSLQRL 360
Query: 344 LENTFRLDLEATREYCIADDKLVNAIEFLDLGDGAGWELLQAMLNADFRKRPTAEAVLSH 403
LENTFRLDLEATREYCIADD+LV+ +EFLDLG+GAGWE+LQAMLNADFRKRPTAEAVL+H
Sbjct: 361 LENTFRLDLEATREYCIADDRLVDGVEFLDLGNGAGWEILQAMLNADFRKRPTAEAVLNH 540
Query: 404 RFMTG 408
RFM+G
Sbjct: 541 RFMSG 555
>AV407455
Length = 399
Score = 113 bits (282), Expect = 7e-26
Identities = 70/127 (55%), Positives = 81/127 (63%), Gaps = 17/127 (13%)
Frame = +3
Query: 26 MGVAAVPLLPS-SIFFTSNKSKLRIKANYGIKF---------ITNSDWFQVGRPIGNYGF 75
M VA VPL P S F S +++ KF I++S+ FQVG+PIG+YGF
Sbjct: 18 MVVAVVPLRPHCSSFHHFTLSSSTPSSHHSPKFRLLRPNATLISDSNSFQVGKPIGSYGF 197
Query: 76 MNVTT-------SSTDQYSLGGGLKTQDVQEGSVKIRLYEGRVSQGPLKQTPVLFKVYPG 128
MNVTT S YS GLKTQDV+EGSVKIRLYEGRVSQGPL+ TPV+FKVYPG
Sbjct: 198 MNVTTVFPSSGPGSEYSYSDFKGLKTQDVEEGSVKIRLYEGRVSQGPLRGTPVVFKVYPG 377
Query: 129 TRAGGVV 135
RA G V
Sbjct: 378 RRAAGAV 398
>TC17140 similar to GB|AAB98753.1|1591472|U67521 C4-dicarboxylate
transporter (mae1) {Methanocaldococcus jannaschii;} ,
partial (5%)
Length = 577
Score = 53.5 bits (127), Expect = 6e-08
Identities = 25/29 (86%), Positives = 27/29 (92%)
Frame = +2
Query: 383 LQAMLNADFRKRPTAEAVLSHRFMTGEVL 411
+QAMLNADFRKRPTAEAVL HRFM+G VL
Sbjct: 275 IQAMLNADFRKRPTAEAVLHHRFMSGAVL 361
>AV426550
Length = 419
Score = 31.6 bits (70), Expect = 0.25
Identities = 22/73 (30%), Positives = 38/73 (51%), Gaps = 6/73 (8%)
Frame = +1
Query: 120 PVLFKVYPGTRAGGVVADMMAANELN------SHMFLQSSSKGISQHLMLLLGGFETTTG 173
PV+ ++ G+R AD++A E++ S + SS+KG+ L+L+L G ++
Sbjct: 169 PVILRLL-GSRVVHDDADILAKREVDPSSEAASASLVDSSAKGLFDRLLLVLHGLLSSYP 345
Query: 174 EQWLAFRDYGKST 186
WL + KST
Sbjct: 346 PCWLRLKPVSKST 384
>AV428120
Length = 346
Score = 29.3 bits (64), Expect = 1.3
Identities = 14/40 (35%), Positives = 20/40 (50%)
Frame = +2
Query: 369 IEFLDLGDGAGWELLQAMLNADFRKRPTAEAVLSHRFMTG 408
++ LD GAGW LL +L +R + L H F+ G
Sbjct: 62 LQILDRNWGAGWHLLSLLLATKSSQRISCLDALKHPFLCG 181
>TC8547 homologue to UP|MMK1_MEDSA (Q07176) Mitogen-activated protein
kinase homolog MMK1 (MAP kinase MSK7) (MAP kinase ERK1)
, partial (71%)
Length = 1098
Score = 28.9 bits (63), Expect = 1.6
Identities = 46/190 (24%), Positives = 72/190 (37%), Gaps = 10/190 (5%)
Frame = +1
Query: 228 LRGLAYMHDHDRLHQSLGPFSVSLNTISESEAPYLIPRLRDLAFSVSVRYSELEDSGPLT 287
LRGL Y+H + LH+ L P ++ LN + L+ F ++ SE +
Sbjct: 166 LRGLKYIHSANVLHRDLKPSNLLLNANCD---------LKICDFGLARVTSETDFMTEYV 318
Query: 288 EGLWARASAASAFTYLEKRAFGIADDIYEAGLLFAYLAFVPFCEAGVMDGLSLQRLL--- 344
W RA L + A D++ G +F L G D + RLL
Sbjct: 319 VTRWYRAPE----LLLNSSDYTAAIDVWSVGCIFMELMDRKPLFPG-RDHVHQLRLLMEL 483
Query: 345 -----ENTFRLDLEATREYC--IADDKLVNAIEFLDLGDGAGWELLQAMLNADFRKRPTA 397
E+ E + Y + + + E +L++ ML D RKR T
Sbjct: 484 IGTPSEDDLGFLNENAKRYIRQLPPYRRQSFQEKFPQVHPEAIDLVEKMLTFDPRKRITV 663
Query: 398 EAVLSHRFMT 407
E L+H ++T
Sbjct: 664 EEALAHPYLT 693
>TC8079 homologue to UP|Q9M6S1 (Q9M6S1) MAP kinase 3, complete
Length = 1550
Score = 27.7 bits (60), Expect = 3.7
Identities = 12/30 (40%), Positives = 19/30 (63%)
Frame = +3
Query: 224 LQGALRGLAYMHDHDRLHQSLGPFSVSLNT 253
L LRGL Y+H + +H+ L P ++ LN+
Sbjct: 612 LYQVLRGLKYIHSANIIHRDLKPSNLLLNS 701
>TC15168 similar to UP|O04084 (O04084) Serine carboxypeptidase isolog,
30227-33069, partial (55%)
Length = 1005
Score = 27.3 bits (59), Expect = 4.8
Identities = 21/62 (33%), Positives = 26/62 (41%)
Frame = +1
Query: 145 NSHMFLQSSSKGISQHLMLLLGGFETTTGEQWLAFRDYGKSTAADYAKVASEKVSKLSSW 204
N M L KGI LLG ET+ E W+ DY S A + + V K +
Sbjct: 61 NKDMSLHIDLKGI------LLGNPETSDAEDWMGLVDYAWSHAV-ISDETHQTVKKSCDF 219
Query: 205 NS 206
NS
Sbjct: 220 NS 225
>TC16135 UP|PSAB_LOTJA (P58385) Photosystem I P700 chlorophyll A apoprotein
A2 (PsaB) (PSI-B), complete
Length = 2647
Score = 27.3 bits (59), Expect = 4.8
Identities = 13/33 (39%), Positives = 19/33 (57%)
Frame = +1
Query: 145 NSHMFLQSSSKGISQHLMLLLGGFETTTGEQWL 177
+SH+F +S+G ++ LLGGF T WL
Sbjct: 718 SSHLF--GTSQGAGTAILTLLGGFHPQTQSLWL 810
>TC17660 homologue to UP|Q9M6R8 (Q9M6R8) MAP kinase PsMAPK2, partial (64%)
Length = 717
Score = 26.9 bits (58), Expect = 6.2
Identities = 11/25 (44%), Positives = 17/25 (68%)
Frame = +2
Query: 228 LRGLAYMHDHDRLHQSLGPFSVSLN 252
LRGL Y+H + LH+ L P ++ +N
Sbjct: 263 LRGLKYLHSANILHRDLKPGNLLIN 337
>AV768437
Length = 393
Score = 26.6 bits (57), Expect = 8.1
Identities = 25/86 (29%), Positives = 41/86 (47%), Gaps = 3/86 (3%)
Frame = +2
Query: 201 LSSWNSFERGQSMKRRRRFIIKLLQGALRGLAYMHDHDR---LHQSLGPFSVSLNTISES 257
LSS N+ + G +R+ I L G RGLAY+H+ LH + P ++ L T +
Sbjct: 89 LSSSNALDWG------KRYNIAL--GTARGLAYLHEECLEWILHCDIKPQNILLGTDYQ- 241
Query: 258 EAPYLIPRLRDLAFSVSVRYSELEDS 283
P++ D S ++ + L +S
Sbjct: 242 ------PKVADFGLSKLLQRNNLNNS 301
>TC11097 similar to UP|P93520 (P93520) Calcium/calmodulin-dependent protein
kinase homolog|CaM kinase homolog|MCK1, partial (36%)
Length = 949
Score = 26.6 bits (57), Expect = 8.1
Identities = 10/26 (38%), Positives = 18/26 (68%)
Frame = +2
Query: 381 ELLQAMLNADFRKRPTAEAVLSHRFM 406
+ ++ +LN D+RKR TA L+H ++
Sbjct: 158 DFVKRLLNKDYRKRMTAAQALTHPWL 235
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.319 0.135 0.391
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,059,621
Number of Sequences: 28460
Number of extensions: 72421
Number of successful extensions: 408
Number of sequences better than 10.0: 24
Number of HSP's better than 10.0 without gapping: 405
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 407
length of query: 411
length of database: 4,897,600
effective HSP length: 92
effective length of query: 319
effective length of database: 2,279,280
effective search space: 727090320
effective search space used: 727090320
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 56 (26.2 bits)
Medicago: description of AC145222.13


